Team:Heidelberg/Project/Capsid Shuffling/ViroBytes
From 2010.igem.org
(→Introduction) |
(→Protocols) |
||
| (14 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
<br /> | <br /> | ||
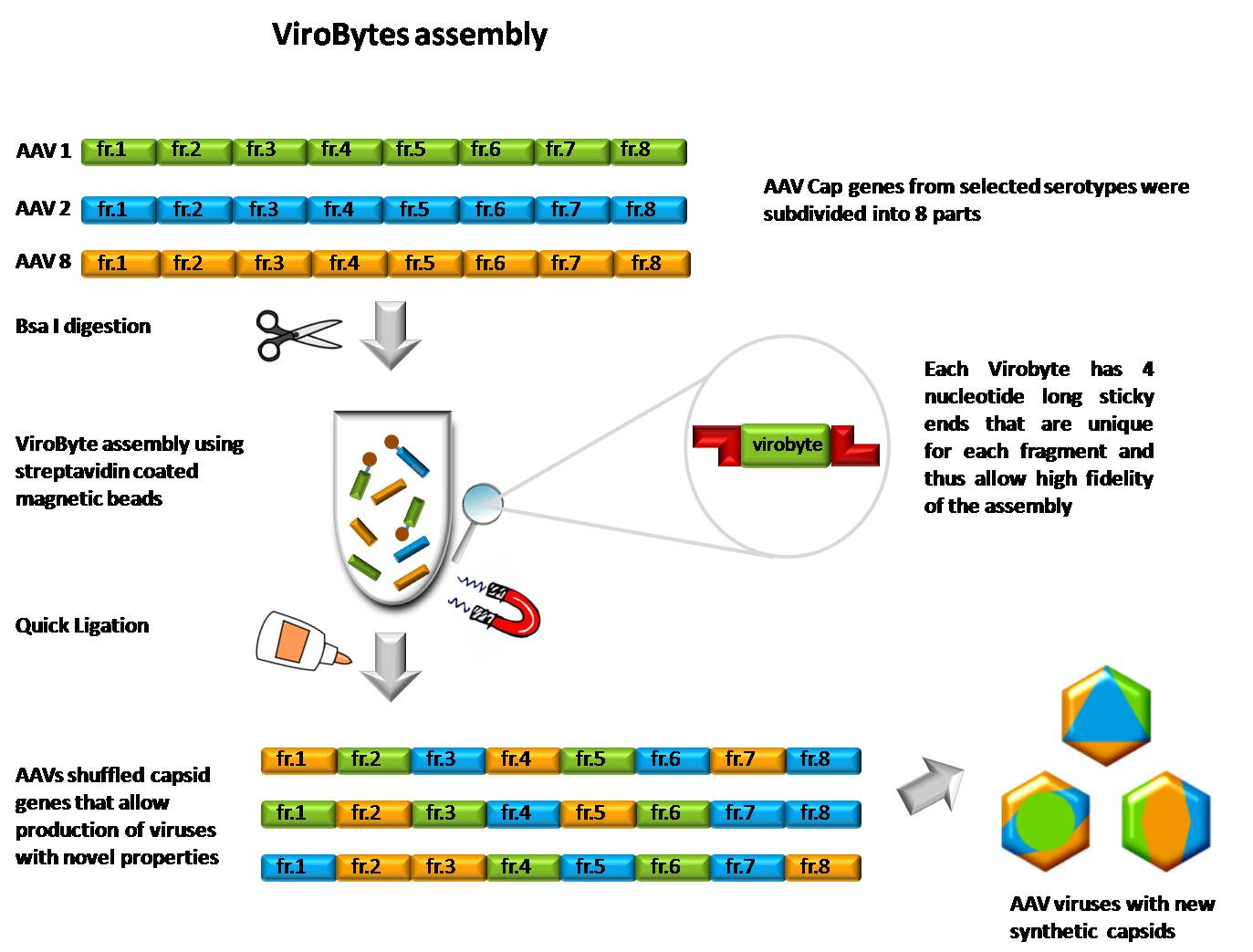
| - | [[Image: | + | [[Image:Virobytes_final_1.jpg|thumb|350px|center|ViroBytes Assembly Scheme]] |
==Important facts== | ==Important facts== | ||
| Line 70: | Line 70: | ||
==ViroByte design== | ==ViroByte design== | ||
| + | <br /> | ||
| - | + | ===Anchor=== | |
| - | 5'- | + | Anchor for ViroByte assembly was designed according to [https://2009.igem.org/Team:Alberta/DNAanchor BioByte Anchor System]. We used 5'-biotinylated oligos which were annealed with Anchor_complementary sequences (see [https://2010.igem.org/Team:Heidelberg/Project/Capsid_Shuffling/ViroBytes#Protocols Protocols]). The final Anchor construct contains important restriction sites as well as the VirByte_for sequence used for final PCR amplification. First recognition site is HindIII which we used for separating the beads from the assembled DNA before PCR. Second restriction sequence corresponds to PacI which is a common non-cutter within all AAV serotypes used and can be emplyed for further ligation into an appropriate vector. Most importantly the 3'-end forms a sticky end compatible with the so-called "start overhang" of all fragments'''1'''. |
| - | + | Annotated sequence of anchor oligos: | |
| - | + | <br /> | |
| - | + | Anchor: | |
| - | - ''''' | + | 5'-Biotin-tctctctctctctct''aagctt'' '''gtctgagtgactagcattcg''' '''''ttaattaa'''''c+4nt(cap specific) |
| - | *Anchor complement: | + | *first 15nt form ssDNA linker (improves efficiency of Anchor binding onto Steptavidin-coated magnetic beads) |
| + | |||
| + | * ''italicized'' is HindIII recognition site | ||
| + | |||
| + | * '''bold''' is amplification (VirByte_for) sequence | ||
| + | |||
| + | * '''''bold italicized''''' is PacI recognition site | ||
| + | |||
| + | Anchor complement: | ||
5'-g'''''ttaattaa''''' '''cgaatgctagtcactcagac''' ''aagctt'' | 5'-g'''''ttaattaa''''' '''cgaatgctagtcactcagac''' ''aagctt'' | ||
| - | + | * reverse complimentary strand to Anchors | |
| - | + | Final Anchor construct used: | |
| + | |||
| + | 5'-Biotin-tctctctctctctctaagcttgtctgagtgactagcattcgttaattaac+4nt-3' | ||
| + | 3'-ttcgaacagactcactgatcgtaagcaattaattg-5' | ||
| + | |||
| + | ===Primers=== | ||
| + | |||
| + | Fragment X forward [https://2010.igem.org/Team:Heidelberg/Notebook/Material/Primer#ViroBytes_primers primers]: | ||
5'-gctac'''ggtctc'''aNNNN+~20-25nt Cap specific | 5'-gctac'''ggtctc'''aNNNN+~20-25nt Cap specific | ||
| - | + | * '''bold''' is BsaI recognition sequence | |
| - | + | * NNNN = cap specific homology sequence from all cap genes (also cutting sequence of BsaI) forming the 5'-overhang | |
| - | + | Fragment X reverse primers: | |
5'-gctac'''ggtctc'''gNNNN | 5'-gctac'''ggtctc'''gNNNN | ||
| - | + | * '''bold''' is BsaI recognition sequence | |
==Protocols== | ==Protocols== | ||
| + | ==='''ViroByte construction'''=== | ||
| + | |||
| + | *perform ViroByte Phusion HiFi Hot Start PCR to obtain desired fragments with complementary ends | ||
| + | **98°C 30s 1x | ||
| + | **98°C 10s | ||
| + | **72°C 45s 35x | ||
| + | **72°C 10min 1x | ||
| + | **4°C hold | ||
| + | |||
| + | |||
| + | *digestion with Bsa1 - leave digesting longer than in standard protocol (at least ~3hrs) | ||
| + | |||
| + | ==='''Anchor preparation'''=== | ||
| + | |||
| + | *Mix 10ul of Anchor 5 (100uM stock) with 50ul of Anchor 12689 (100ul stock); add 1.08ml of nuclease free water; mix gently | ||
| + | |||
| + | *heat on the block to 95degrees for 2 minutes and let cool slowly to room temperature | ||
| + | |||
| + | ==='''Anchor docking'''=== | ||
| + | |||
| + | *after washing the beads and aspiration of the cleared solution the beads are ready for adding of the Anchor | ||
| + | |||
| + | *add 40ul of the Anchor solution (~200ng/ul) | ||
| + | |||
| + | *resuspend the beads and let incubate at room temperature for 15-20 minutes with occasional flicking in order to maintain the homogeneity of the solution | ||
| + | |||
| + | *applying magnet, wash twice with 80ul of wash/binding buffer (0.5 M NaCl; 20 mM Tris HCl (pH 7.5); 1 mM EDTA) and once with 80ul of 1X T4 ligase buffer | ||
| + | |||
| + | ==='''BioByte-like assembly'''=== | ||
| + | |||
| + | *Bead preparation | ||
| + | |||
| + | *Quickly vortex beads stock solution to obtain homogeneous suspension | ||
| + | |||
| + | *Aliquot 40 µl of the beads into 1.5ml Eppendorf tubes - gently tap the tubes to collect all the beads at the bottom. | ||
| + | |||
| + | *Place the tubes on the magnetic rack (or place a magnet on the side of each tube) and wait for the beads to be cleared out from the solution | ||
| + | |||
| + | *Carefully discard the supernatant whilst beads are still attached to the side wall | ||
| + | |||
| + | *Resuspend beads in 80ul of wash/binding buffer by gentle flicking (vortexing at high speed is not recommended) | ||
| + | |||
| + | *Wash the beads once with 80 µl of 1X T4 Ligase buffer, before addition of ligation premix. | ||
| + | |||
| + | *Add the DNA fragments using the ligation premix: 1) 25 µl of Quick Ligase (2X buffer); 2) Amount of DNA corresponding to 200 ng. The optimal DNA concentration for the ViroByte fragments assembly was experimentally established and was in range of 200 ng per fragment. Using higher and lower DNA concentrations did not give any positive assembly results; 3) Fill up with water to obtain the 50 µl total volume. | ||
| + | |||
| + | *For the ligation procedure the Quick Ligase was used that showed to be more efficient during the experimental procedure in comparison to the ordinary T4 Ligase. Optimal ligation time for the Virobytes ligation was between 15-25 minutes. | ||
| + | |||
| + | *After the last DNA fragment ligation wash the beads once with Wash/Binding buffer. Moreover the DNA should be cleaved from the beads by using HindIII restriction enzyme (or the other one depending on your designed cleavage site). For the restriction a standard protocol for the DNA digestion should be used. | ||
| - | + | ==Notebook== | |
| + | For further reference please find our lab notebook [https://2010.igem.org/Team:Heidelberg/Notebook/Capsid_Shuffling/ViroBytes here]. | ||
==References== | ==References== | ||
Latest revision as of 03:57, 28 October 2010

|
|
|||
 "
"