Team:Groningen/Expression
From 2010.igem.org
(→Subtilin induced expression of chaplins) |
(→Subtilin induced expression of chaplins) |
||
| (50 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
===Subtilin induced expression of chaplins=== | ===Subtilin induced expression of chaplins=== | ||
| - | |||
The biofilm forming capacity of ''Bacillus subtilis'' makes it a good host for our application. In addition, ''B. subtilis'' is known for its ability to produce and secrete large amounts of protein at high cell densities. However, despite its track record as an efficient production organism and the fact that both ''B. subtilis'' and ''Streptomyces coelicolor'' are gram-positive bacteria, it is not certain wether chaplins can be heterologously expressed in ''B. subtilis''. Improper folding, unsuccessful export, or even the very nature of the chaplins, could still lead to hampered expression. | The biofilm forming capacity of ''Bacillus subtilis'' makes it a good host for our application. In addition, ''B. subtilis'' is known for its ability to produce and secrete large amounts of protein at high cell densities. However, despite its track record as an efficient production organism and the fact that both ''B. subtilis'' and ''Streptomyces coelicolor'' are gram-positive bacteria, it is not certain wether chaplins can be heterologously expressed in ''B. subtilis''. Improper folding, unsuccessful export, or even the very nature of the chaplins, could still lead to hampered expression. | ||
We took several steps to ensure optimal expression. The coding sequences of the chaplins were codon optimized for ''B. subtilis'' and synthesized. We placed a ribosome binding site in front of the coding sequences that is known to work well in ''B. subtilis'', and flanked these constructs with the biobrick prefix and suffix. | We took several steps to ensure optimal expression. The coding sequences of the chaplins were codon optimized for ''B. subtilis'' and synthesized. We placed a ribosome binding site in front of the coding sequences that is known to work well in ''B. subtilis'', and flanked these constructs with the biobrick prefix and suffix. | ||
| Line 17: | Line 16: | ||
<br> | <br> | ||
[[Image:SURE-gfp-gn.jpg|250px|thumb|right|Subtilin induction of GFP by the SURE system (Bongers ''et al'', 2005)]] | [[Image:SURE-gfp-gn.jpg|250px|thumb|right|Subtilin induction of GFP by the SURE system (Bongers ''et al'', 2005)]] | ||
| - | Because it is uncertain how chaplin expression will affect ''B. subtilis'', the initial expression attempts were performed with the stringently controlled, subtilin-regulated gene expression (SURE) system (Bongers ''et al'', 2005). This system uses the subtilin sensing machinery present in a strain of ''B. subtilis'' that autoinduces the production of more of the [http://en.wikipedia.org/wiki/Lantibiotics lantibiotic] subtilin. The subtilin sensor histidine kinase SpaK phosphorylates the response regulator SpaR, which can then bind to so-called ''spa'' boxes in the promoter regions of genes involved in subtilin biosynthesis (Kleerebezem ''et al'', 2004). In the SURE system, a ''B. subtilis'' strain naturally lacking the subtilin biosynthesis genes has the ''spaRK'' genes introduced into its genome. A plasmid carrying a ''spa'' box promoter that is transformed to this strain can then drive the expression of proteins upon subtilin induction of SpaRK signalling. | + | Because it is uncertain how chaplin expression will affect ''B. subtilis'', the initial expression attempts were performed with the stringently controlled, subtilin-regulated gene expression (SURE) system (Bongers ''et al'', 2005). This system uses the subtilin sensing machinery present in a strain of ''B. subtilis'' that autoinduces the production of more of the [http://en.wikipedia.org/wiki/Lantibiotics lantibiotic] subtilin. The subtilin sensor histidine kinase SpaK phosphorylates the response regulator SpaR, which can then bind to so-called ''spa'' boxes in the promoter regions of genes involved in subtilin biosynthesis (Kleerebezem ''et al'', 2004). In the SURE system, a ''B. subtilis'' strain naturally lacking the subtilin biosynthesis and regulation genes has the ''spaRK'' genes introduced into its genome. A plasmid carrying a ''spa'' box promoter that is transformed to this strain can then drive the expression of proteins upon subtilin induction of SpaRK signalling. |
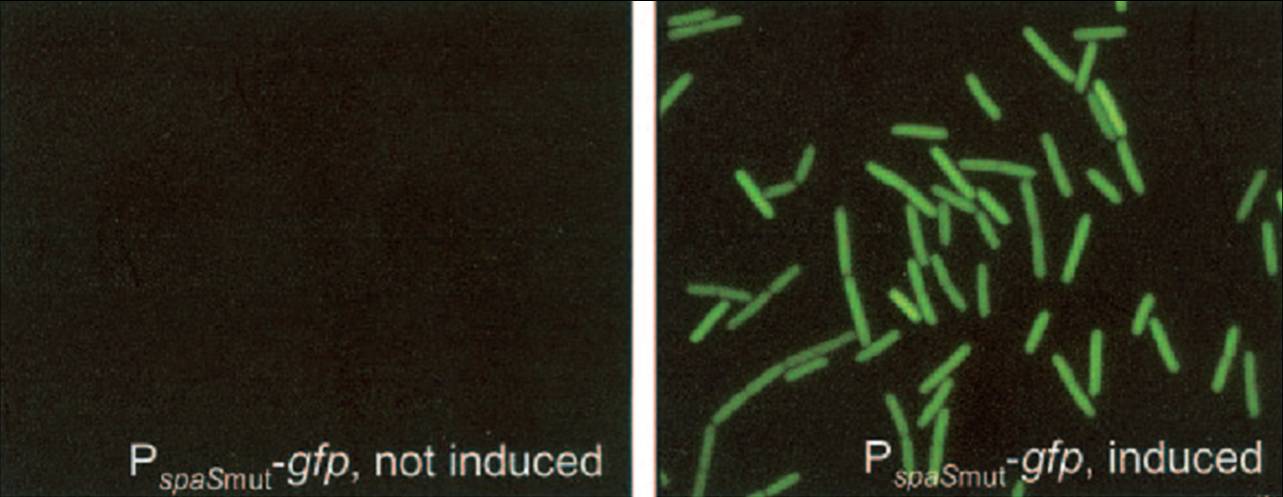
| - | [[Image: | + | [[Image:Sub-ind-gn.jpg|250px|thumb|right|Subtilin induced GFP expression in Bacillus subtilis using the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305011 BBa_K305011] backbone]] |
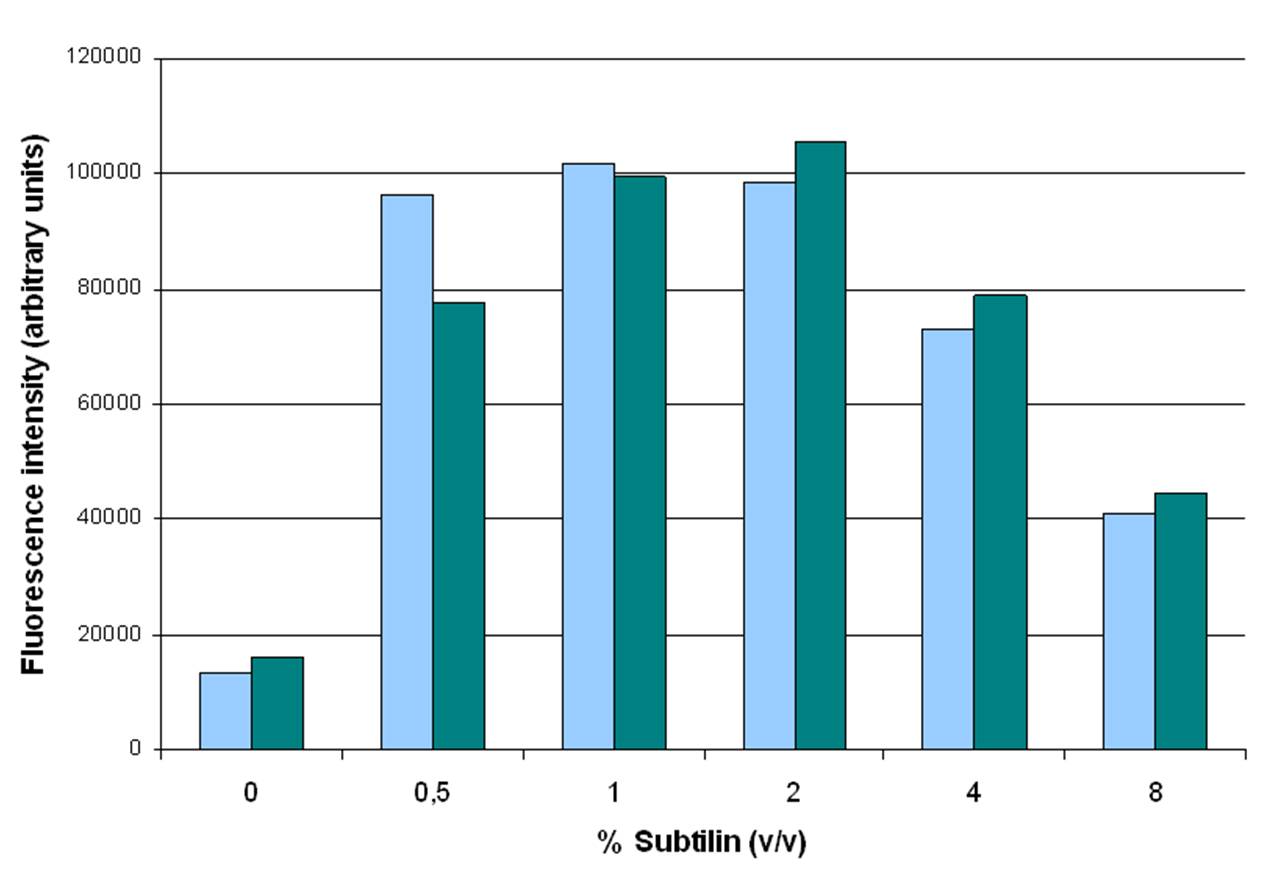
| - | We have adapted this system to make it BioBrick compatible for easy expression of our chaplins, combinations of chaplins, or any other biobrick part that is composed of an RBS followed by a protein coding sequence. We introduced the BioBrick prefix and suffix into the expression plasmid, downstream of the mutated ''spaS'' promoter, producing our subtilin inducible expression backbone part, [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305011 BBa_K305011]. To test the expression and find a suitable subtilin concentration for induction of the chaplins we made use of GFP fluorescence measurements. We inserted the part [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0240 BBa_E0240] into the BioBrick site and induced liquid cultures of ''B. subtilis'' carrying this plasmid (and the ''spaRK'' genes) with different volumes of subtilin-containing culture supernatant of a subtilin producing strain of ''B. subtilis''. These results demonstrate that addition of 0.5 to 1%(vol/vol) of subtilin to the culture is sufficient to reach optimal induction. | + | We have adapted this system to make it BioBrick compatible for easy expression of our chaplins, combinations of chaplins, or any other biobrick part that is composed of an RBS followed by a protein coding sequence. We introduced the BioBrick prefix and suffix into the expression plasmid, downstream of the mutated ''spaS'' promoter, producing our subtilin inducible expression backbone part, [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305011 BBa_K305011]. To test the expression and find a suitable subtilin concentration for induction of the chaplins we made use of GFP fluorescence measurements. We inserted the part [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0240 BBa_E0240] into the BioBrick site and induced liquid cultures of ''B. subtilis'' carrying this plasmid (and the ''spaRK'' genes) with different volumes of subtilin-containing culture supernatant of a subtilin producing strain of ''B. subtilis''. These results demonstrate that addition of 0.5 to 1%(vol/vol) of subtilin to the culture is sufficient to reach optimal induction. |
| + | <br> | ||
'''Chaplin detection''' | '''Chaplin detection''' | ||
| + | <br> | ||
| + | In our first expression tests we focused on detecting the chaplins in either the medium in which our chaplin producing population grows | ||
| + | <br> | ||
| + | Using the same methods that were used to dissolve and monomerize chaplins from Streptomyces, we treated cell pellets and [http://en.wikipedia.org/wiki/Trichloricacetic_acid TCA] precipitated supernatant with [http://en.wikipedia.org/wiki/Trifluoroacetic_acid TFA] to purify chaplin proteins. [http://en.wikipedia.org/wiki/Trifluoroacetic_acid TFA] treatment with 99% pure TFA breaks down most proteins and monomerises assembled chaplin fibers, this enables us to detect the chaplins on SDS gel. | ||
| - | + | <br> | |
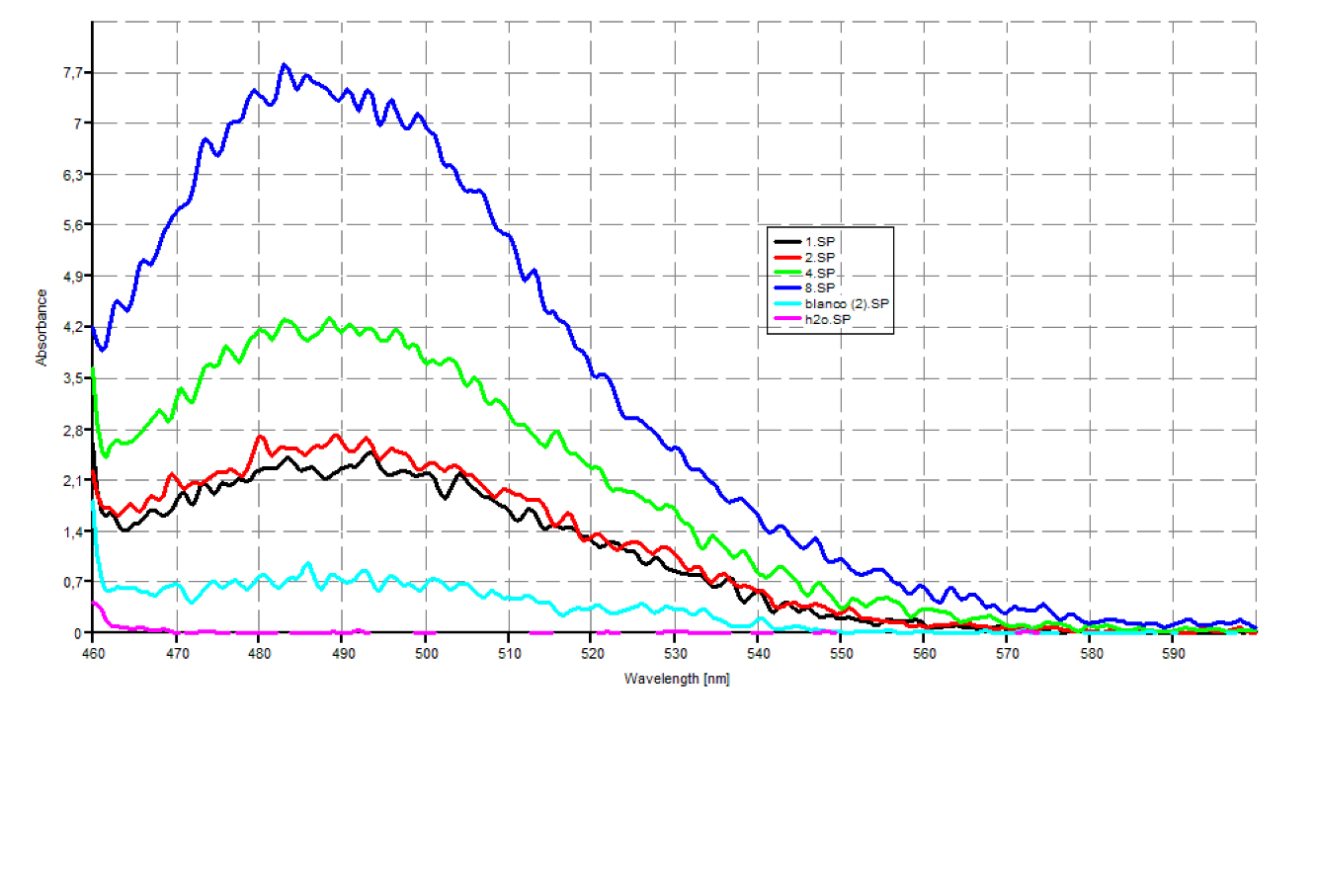
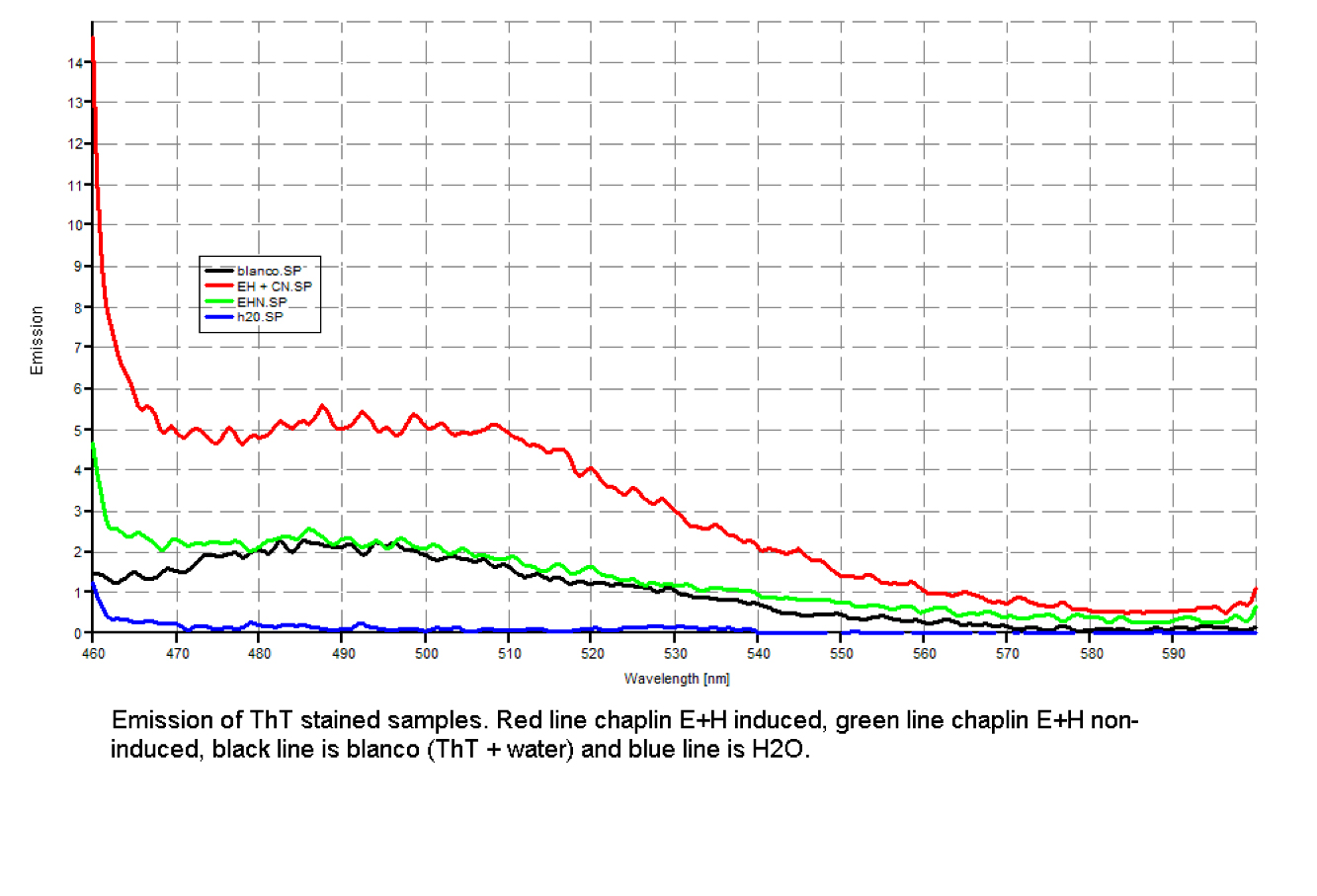
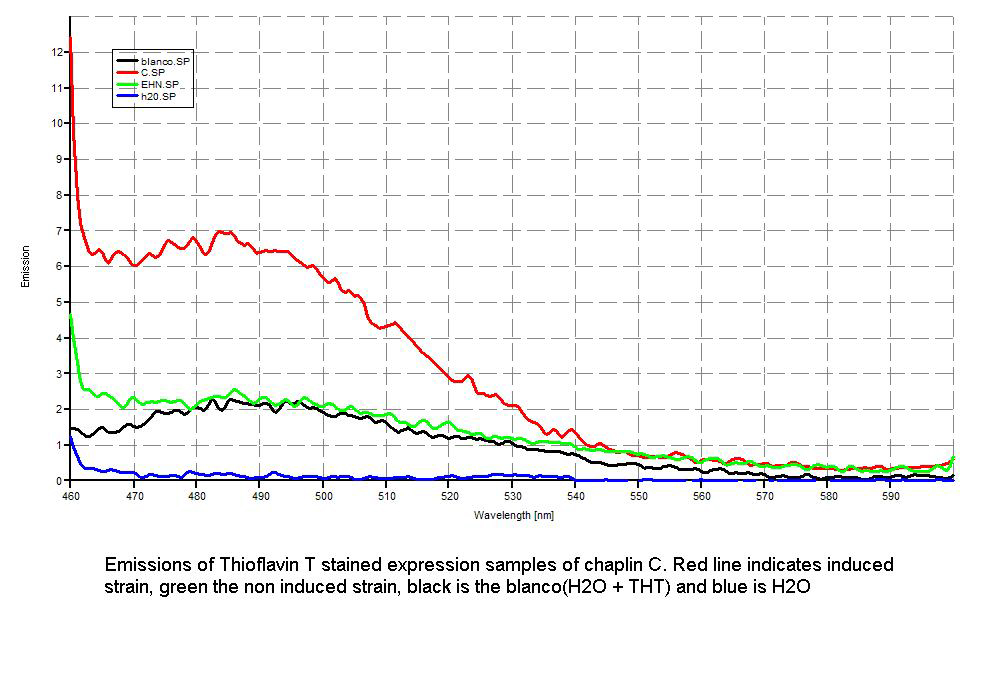
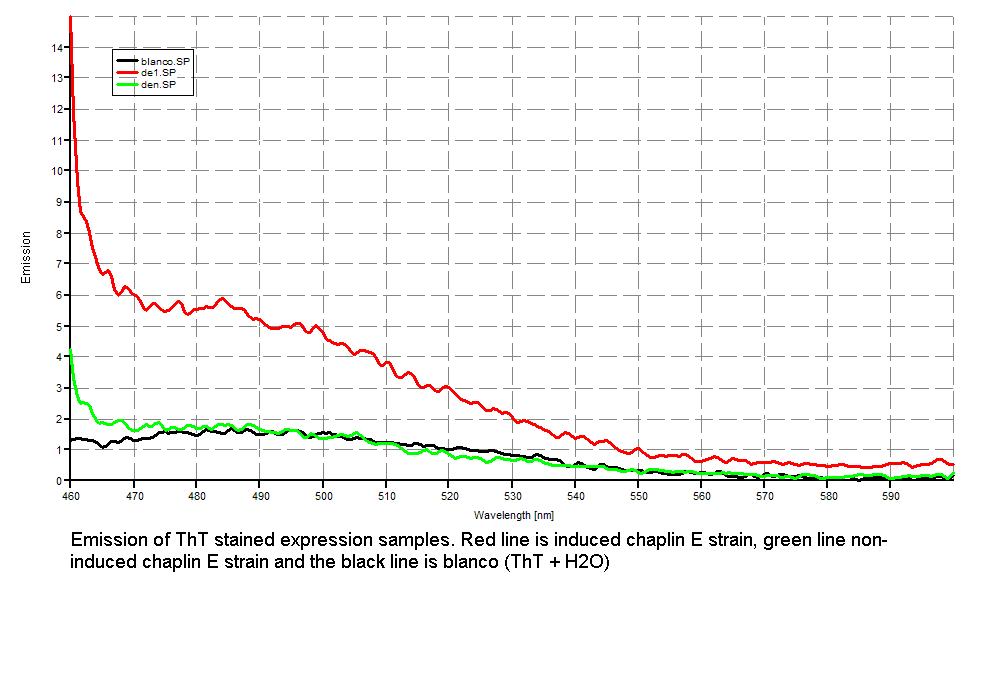
| - | + | Since our early expression experiments didn't yield conclusive results regarding the detection of our chaplins, we tried staining our samples with an amyloid specific stain called [http://en.wikipedia.org/wiki/Thioflavin Thioflavin T]. Initial testing with the supernatant and washed pellet gave [intriguing resultsGr] yet not clear. Our emission graphs showed some irregularities with the subtilin induced samples, but seemed to be distorted by background noise caused by other materials in the sample. To further purify our samples we decided to [extractioncellwallsGR disrupt] our liquid culture and boil it two times in 2% SDS, before treating the freeze dried sample with 99%. TFA Using such a harsh method, we hope to denaturate most proteins to prevent their interference in chaplin detection and highten the relative concentration of chaplin proteins in tested samples, this turned out to be a more successful method. | |
| + | |||
| + | Using this method we succesfully detected chaplins C, E and H in purified cell walls from induced B. subtilis cultures, confirmation of these results was provided by malditov mass spectrometry. | ||
| + | <br> | ||
| + | <html> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td></html>[[Image:Chgr.jpg|300px|thumb|Purified chaplins from S. coelicolor exitate at 482 when stained with THT. Different dilutions were used to set up a ladder.]]<html></td> | ||
| + | <td></html>[[Image:EHgr.jpg|300px|thumb|Purified E and H chaplins from B subtilis exitate at 482 when stained with THT]] <html></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></html>[[Image:Cgr.jpg|300px|thumb|Purified C chaplins from B subtilis exitate at 482 when stained with THT]]<html></td> | ||
| + | <td></html>[[Image:Egr.jpg|300px|thumb|Purified E chaplins from B subtilis exitate at 482 when stained with THT]]<html></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
| + | <br> | ||
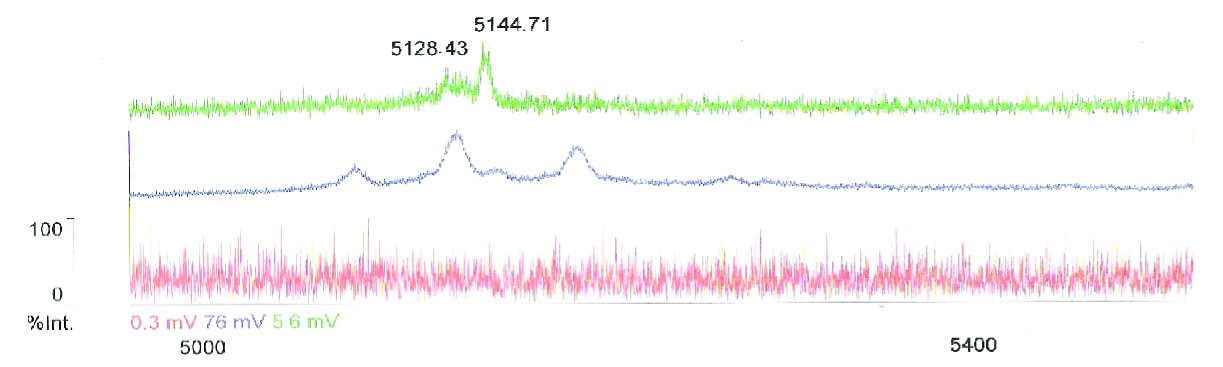
| + | [[Image:MassSpecGR.jpg|700px|thumb|none|Mass spectometry results confirm the production of Chaplin H (5128) and E (5144) in Bacillus (green). Peaks is Streptomyces (blue) corrispond, although the peak of chaplin E (5144) indicates that somewhere during translation or in post-translational processes a 21kb threonine group is lost. The non induced B. subtilis strain serves as a control (red). Chaplin C has not been detected this way, but that was not to be expected (Claessen et al. (2009).]] | ||
===Timed expression of chaplins in a biofilm === | ===Timed expression of chaplins in a biofilm === | ||
| Line 33: | Line 54: | ||
[[Image:Groningen-Promotors-sketch.png|300px|left]] | [[Image:Groningen-Promotors-sketch.png|300px|left]] | ||
| - | |||
| - | |||
'''''srfA''''' | '''''srfA''''' | ||
| + | <br> | ||
| + | |||
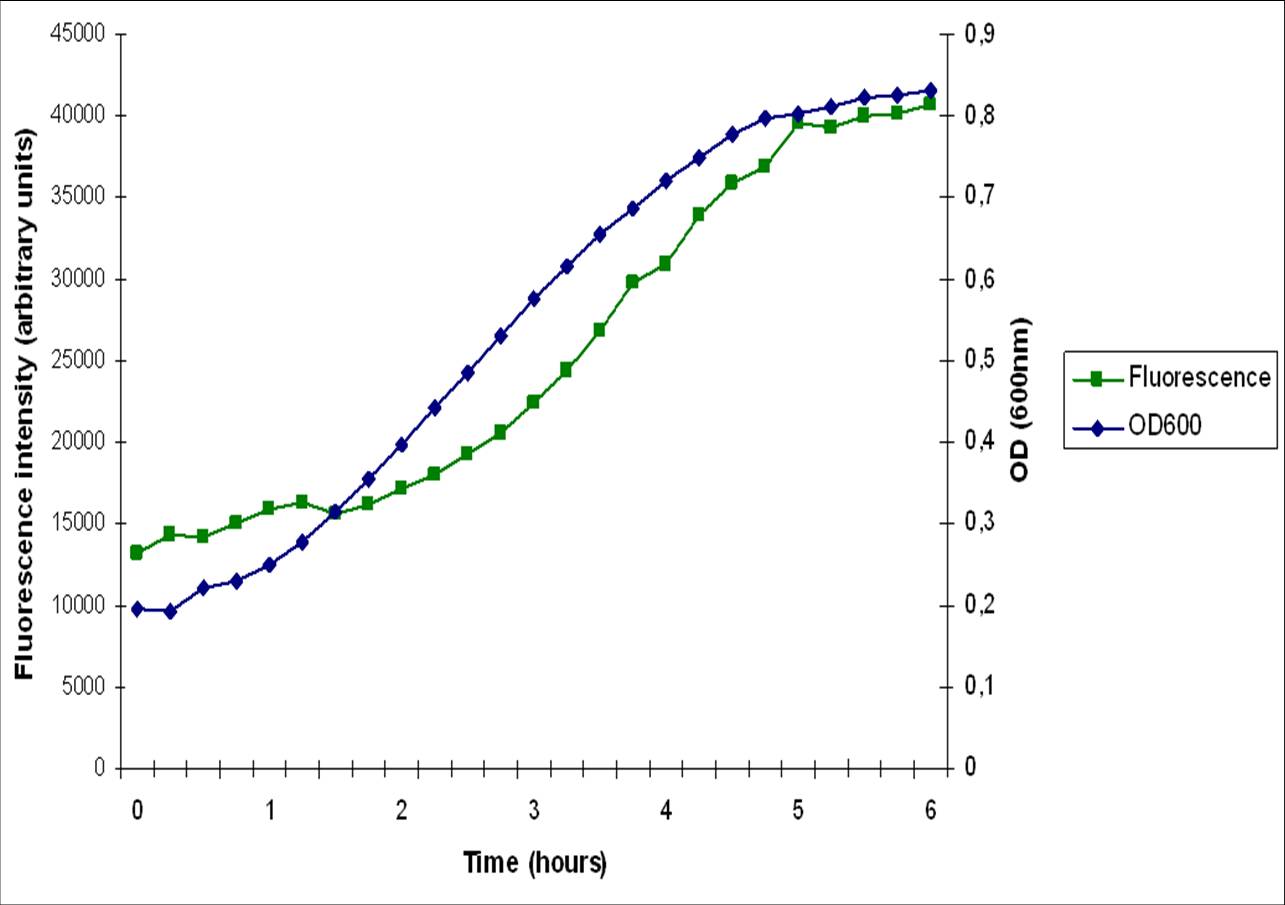
| + | The [http://dbtbs.hgc.jp/COG/prom/srfAA-srfAB-comS-srfAC-srfAD.html ''srfA'' operon] has been reported to be important for natural competence and sporulation in ''Bacillus subtilis''. All these activities occur in biofilms, the promoter is not active until the end of exponential growth. It is controlled by the [https://2010.igem.org/Team:Groningen/Expression_model#ComXPA_quorum_sensing_system ComXPA quorum sensing system] and hence active in states of high cell densities. Therefore the ''srfA'' promoter would be suitable for chaplin expression. Two different lengths of the ''srfA'' promoter where chosen due to uncertainties concerning the region between the response element and the transcription start side of the SrfAA protein. In the original promoter this region is unusually long, by shortening it 190bp’s we hope to achieve a higher transcription efficiency. So we came up with two different promoters, the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305008 original] one and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305007 shortened] one. We followed the promoter activity by placing a RBS+GFP BioBrick downstream of these promoters. We succesfully demonstrated that the short ''srfA''-promoter variant drives the expression of GFP in a cell density dependent way. However, as can be seen in the image below, the fluorescence starts increasing during early exponential growth, rather than late exponential growth. This might be because this multi-copy plasmid system is more sensitive to srfA promoter activators. It could also be caused by our removal of the sequences downstream of the regulator binding sites and upstream of the srfA coding sequence. These sequences may have a regulatory role, preventing transcription/translation at earlier growth stages. | ||
| + | |||
| - | + | [[image:igemgroningen_srfa_Promotoractivity.jpg|left|200px|srfA|thumb|srfA promotor activity during cell growth (Nakano MM. 1991)]] | |
| - | + | [[Image:florescence_srfA.jpg|left|300px|thumb|Promoter activity of our short srfaA promoter determined by measuring GFP reporter fluorescence, with overlaid growth curve.]] | |
| + | <br style="clear: both" /> | ||
Latest revision as of 03:58, 28 October 2010
Expression of chaplins
Summary
The goal of our project is to let Bacillus subtilis make a hydrophobic coating by forming a biofilm and then expressing and secreting chaplins. However, first we needed to test whether B. subtilis was capable of expressing chaplins, since they could impair the cellgrowth due to their hydrophobic and self assembling properties. We succesfully expressed chaplins C, E and H in B. subtilis using a tightly regulated subtilin inducable system called "SURE". Furthermore we tested the SURE system for optimal subtilin concentration with GFP. We want B. subtilis to auto-induce the expression of the chaplins after biofilmformation. Therefore we looked into two operons in B. subtilis; one that gets triggered in late exponential growth (srfA operon) and one that is involved in the formation of biofilm (yqxM-sipW-tasA operon). Using the srfA promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K305007 BBa_K305007]), we succesfully expressed GFP demonstrating that this promoter could be used to auto-induce the expression chaplins.
Subtilin induced expression of chaplins
The biofilm forming capacity of Bacillus subtilis makes it a good host for our application. In addition, B. subtilis is known for its ability to produce and secrete large amounts of protein at high cell densities. However, despite its track record as an efficient production organism and the fact that both B. subtilis and Streptomyces coelicolor are gram-positive bacteria, it is not certain wether chaplins can be heterologously expressed in B. subtilis. Improper folding, unsuccessful export, or even the very nature of the chaplins, could still lead to hampered expression. We took several steps to ensure optimal expression. The coding sequences of the chaplins were codon optimized for B. subtilis and synthesized. We placed a ribosome binding site in front of the coding sequences that is known to work well in B. subtilis, and flanked these constructs with the biobrick prefix and suffix.
SURE expression system
Because it is uncertain how chaplin expression will affect B. subtilis, the initial expression attempts were performed with the stringently controlled, subtilin-regulated gene expression (SURE) system (Bongers et al, 2005). This system uses the subtilin sensing machinery present in a strain of B. subtilis that autoinduces the production of more of the [http://en.wikipedia.org/wiki/Lantibiotics lantibiotic] subtilin. The subtilin sensor histidine kinase SpaK phosphorylates the response regulator SpaR, which can then bind to so-called spa boxes in the promoter regions of genes involved in subtilin biosynthesis (Kleerebezem et al, 2004). In the SURE system, a B. subtilis strain naturally lacking the subtilin biosynthesis and regulation genes has the spaRK genes introduced into its genome. A plasmid carrying a spa box promoter that is transformed to this strain can then drive the expression of proteins upon subtilin induction of SpaRK signalling.
We have adapted this system to make it BioBrick compatible for easy expression of our chaplins, combinations of chaplins, or any other biobrick part that is composed of an RBS followed by a protein coding sequence. We introduced the BioBrick prefix and suffix into the expression plasmid, downstream of the mutated spaS promoter, producing our subtilin inducible expression backbone part, [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305011 BBa_K305011]. To test the expression and find a suitable subtilin concentration for induction of the chaplins we made use of GFP fluorescence measurements. We inserted the part [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0240 BBa_E0240] into the BioBrick site and induced liquid cultures of B. subtilis carrying this plasmid (and the spaRK genes) with different volumes of subtilin-containing culture supernatant of a subtilin producing strain of B. subtilis. These results demonstrate that addition of 0.5 to 1%(vol/vol) of subtilin to the culture is sufficient to reach optimal induction.
Chaplin detection
In our first expression tests we focused on detecting the chaplins in either the medium in which our chaplin producing population grows
Using the same methods that were used to dissolve and monomerize chaplins from Streptomyces, we treated cell pellets and [http://en.wikipedia.org/wiki/Trichloricacetic_acid TCA] precipitated supernatant with [http://en.wikipedia.org/wiki/Trifluoroacetic_acid TFA] to purify chaplin proteins. [http://en.wikipedia.org/wiki/Trifluoroacetic_acid TFA] treatment with 99% pure TFA breaks down most proteins and monomerises assembled chaplin fibers, this enables us to detect the chaplins on SDS gel.
Since our early expression experiments didn't yield conclusive results regarding the detection of our chaplins, we tried staining our samples with an amyloid specific stain called [http://en.wikipedia.org/wiki/Thioflavin Thioflavin T]. Initial testing with the supernatant and washed pellet gave [intriguing resultsGr] yet not clear. Our emission graphs showed some irregularities with the subtilin induced samples, but seemed to be distorted by background noise caused by other materials in the sample. To further purify our samples we decided to [extractioncellwallsGR disrupt] our liquid culture and boil it two times in 2% SDS, before treating the freeze dried sample with 99%. TFA Using such a harsh method, we hope to denaturate most proteins to prevent their interference in chaplin detection and highten the relative concentration of chaplin proteins in tested samples, this turned out to be a more successful method.
Using this method we succesfully detected chaplins C, E and H in purified cell walls from induced B. subtilis cultures, confirmation of these results was provided by malditov mass spectrometry.
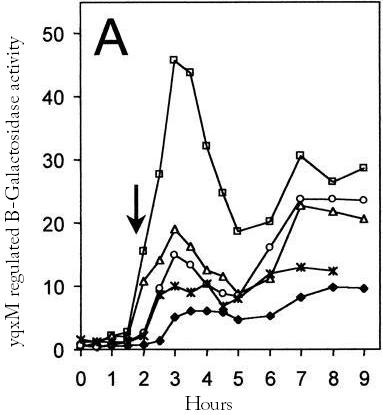
Timed expression of chaplins in a biofilm
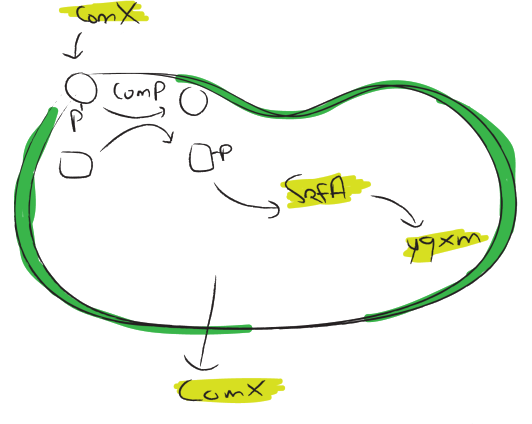
An important question is which promoter we should use to control the chaplin expression. We assume that an ideal promoter would not be active until the biofilm has formed because the expression of hydrophobic proteins might influence the formation of it. Two promoters where found that are active in biofilms but not during normal growth.
srfA
The [http://dbtbs.hgc.jp/COG/prom/srfAA-srfAB-comS-srfAC-srfAD.html srfA operon] has been reported to be important for natural competence and sporulation in Bacillus subtilis. All these activities occur in biofilms, the promoter is not active until the end of exponential growth. It is controlled by the ComXPA quorum sensing system and hence active in states of high cell densities. Therefore the srfA promoter would be suitable for chaplin expression. Two different lengths of the srfA promoter where chosen due to uncertainties concerning the region between the response element and the transcription start side of the SrfAA protein. In the original promoter this region is unusually long, by shortening it 190bp’s we hope to achieve a higher transcription efficiency. So we came up with two different promoters, the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305008 original] one and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305007 shortened] one. We followed the promoter activity by placing a RBS+GFP BioBrick downstream of these promoters. We succesfully demonstrated that the short srfA-promoter variant drives the expression of GFP in a cell density dependent way. However, as can be seen in the image below, the fluorescence starts increasing during early exponential growth, rather than late exponential growth. This might be because this multi-copy plasmid system is more sensitive to srfA promoter activators. It could also be caused by our removal of the sequences downstream of the regulator binding sites and upstream of the srfA coding sequence. These sequences may have a regulatory role, preventing transcription/translation at earlier growth stages.
yqxM
The [http://dbtbs.hgc.jp/COG/prom/yqxM-sipW-tasA.html yqxM-sipW-tasA] operon is controlled by the yqxM promoter. It is needed for biofilm formation because TasA is a key protein of the extracellular matrix. The promotor gets activated via a cascade of other regulatory elements, including SrfA, in response to quorum sensing. Since the chaplins should work in a similar way to TasA we think the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K305006 yqxM] promoter would be very suitable for chaplin expression during the stationary phase. We fused the yqxM promoter with GFP but could not observe any expression, since the GFP worked with the srfA promoter we conclude that the yqxM promoter does not work.
References
Bongers RS, Veening JW, Van Wieringen M, Kuipers OP, and Kleerebezem M. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. [http://aem.asm.org/cgi/content/short/71/12/8818Appl Environ Microbiol 2005 Dec; 71(12) 8818-24. pmid:16332878]
Kleerebezem, M., R. Bongers, G. Rutten, W. M. de Vos, and O. P. Kuipers. 2004. Autoregulation of subtilin biosynthesis in Bacillus subtilis: the role of the spa-box in subtilin-responsive promoters. [http://gbb.eldoc.ub.rug.nl/FILES/root/2004/PeptidesKleerebezem/2004PeptidesKleerebezem.pdf Peptides 25:1415–1424]
Stöver AG, Driks A. Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA. [http://jb.asm.org/cgi/content/short/181/17/5476 J. Bacteriol. Sept. 1999, p. 5476-5481, Vol. 181, No. 17]
Nakano MM, Xia LA, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC208261/ PMC208261]
Frances Chu, Daniel B. Kearns, Anna McLoon, Yunrong Chai, Roberto Kolter and Richard Losicka, A Novel Regulatory Protein Governing Biofilm Formation in Bacillus subtilis [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2430766/ PMC2430766]
Hayashi K, Ohsawa T, Kobayashi K, Ogasawara N, Ogura M. The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1251593/ PMC1251593]
 "
"