Team:ETHZ Basel/Biology/Archeal Light Receptor
From 2010.igem.org
(→Receptor analysis) |
|||
| (2 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
As membrane proteins are difficult to express at the appropriate concentrations, we decided to subclone the receptor into the '''IPTG-inducible plasmid pACT3''' allowing us to adapt the expression level. | As membrane proteins are difficult to express at the appropriate concentrations, we decided to subclone the receptor into the '''IPTG-inducible plasmid pACT3''' allowing us to adapt the expression level. | ||
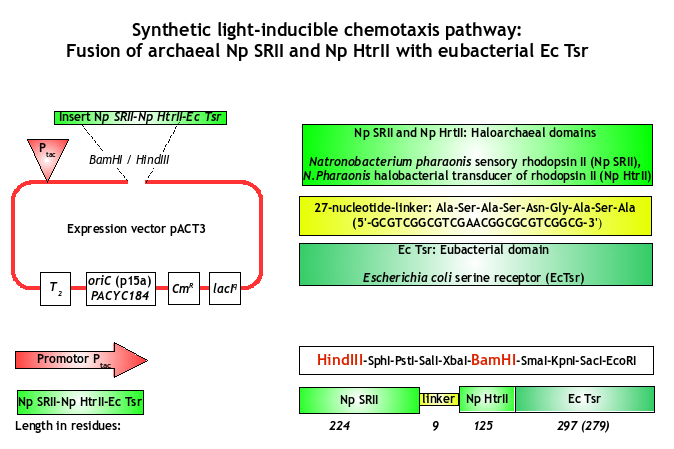
| - | The archeal receptor fusion was cloned into pACT3 via <font color=#ba0000>BamHI</font> and <font color=#ba0000>HindIII</font>. In addition to the BamHI site, the forward primer encodes for a ribosome binding site | + | The archeal receptor fusion was cloned into pACT3 via <font color=#ba0000>BamHI</font> and <font color=#ba0000>HindIII</font>. In addition to the BamHI site, the forward primer encodes for a <font color=#ba0000>ribosome binding site</font> to ensure the optimal spacing to the start codon. |
Forward (5'-3'): GT<font color=#ba0000>GGATCC</font>A<font color=#ba0000>AGGAGA</font>TATACATATGGTTGGTCTGACCACCCTG | Forward (5'-3'): GT<font color=#ba0000>GGATCC</font>A<font color=#ba0000>AGGAGA</font>TATACATATGGTTGGTCTGACCACCCTG | ||
| Line 30: | Line 30: | ||
=Receptor analysis= | =Receptor analysis= | ||
[[Image:AR expression.png|thumb|400px|'''Crude cell extract on a 10 % SDS PAGE.''' '''Legend:''' ''M'' Marker lane; ''Lane 1'' wild type strain; ''Lane 2'' Strain expressing archeal receptor, 0 µM IPTG; ''Lane 3'' Strain expressing archeal receptor, 10 µM IPTG; ''Lane 4'' Strain expressing archeal receptor, 100 µM IPTG; ''Lane 5'' Strain expressing archeal receptor, 1'000 µM IPTG.]] | [[Image:AR expression.png|thumb|400px|'''Crude cell extract on a 10 % SDS PAGE.''' '''Legend:''' ''M'' Marker lane; ''Lane 1'' wild type strain; ''Lane 2'' Strain expressing archeal receptor, 0 µM IPTG; ''Lane 3'' Strain expressing archeal receptor, 10 µM IPTG; ''Lane 4'' Strain expressing archeal receptor, 100 µM IPTG; ''Lane 5'' Strain expressing archeal receptor, 1'000 µM IPTG.]] | ||
| - | To detect expression of the | + | To detect expression of the archeal light receptor, we carried out '''expression experiments''' with different IPTG inducer concentrations (0, 10, 100, 1'000 µM). We then loaded the crude cell extract on a 10 % SDS PAGE. Below you find the gel picture. At a fully induced condition, a band at approximately 68 KDa becomes more prominent. We assume this is the expressed archeal light receptor. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | Due to the fact that this gel picture shows the whole protein fraction of an ''E. coli'' cell, we are planning to subclone ([http://partsregistry.org/Part:BBa_K422001 BBa_K422001]) into an expression vector containing a '''his-tag''' for protein purification. | ||
| + | An important experiment that we are planning to do before the Jamboree is to detect the insertion of the archeal light receptor into the '''membrane'''. For that, cells will have to be lysed after an induction phase and the membrane needs to be extracted by fractionated centrifugation. Using a monochromator, absorption of the membrane fraction at 500 nm (blue light) can be evaluated and compared between wild type and archeal light receptor expressing strain. | ||
= References = | = References = | ||
[1] [http://jb.asm.org/cgi/content/full/183/21/6365?maxtoshow=&hits=10&RESULTFORMAT=1&andorexacttitle=and&andorexacttitleabs=and&fulltext=An+archaeal+photosignal-transducing+module+mediates+phototaxis+in+%27%27Escherichia+&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&resourcetype=HWCIT: Jung, Spudich, Trivedi and Spudich: An archaeal photosignal-transducing module mediates phototaxis in ''Escherichia coli''. Journal of bacteriology. 2001; 21]. | [1] [http://jb.asm.org/cgi/content/full/183/21/6365?maxtoshow=&hits=10&RESULTFORMAT=1&andorexacttitle=and&andorexacttitleabs=and&fulltext=An+archaeal+photosignal-transducing+module+mediates+phototaxis+in+%27%27Escherichia+&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&resourcetype=HWCIT: Jung, Spudich, Trivedi and Spudich: An archaeal photosignal-transducing module mediates phototaxis in ''Escherichia coli''. Journal of bacteriology. 2001; 21]. | ||
Latest revision as of 21:07, 27 October 2010
Archeal Light Receptor
Besides the light-sensitive Pif3/PhyB-system, another implementation strategy caught our attention: The generation of our E. lemming by the fusion of an archeal photoreceptor to a bacterial chemotactic transducer. This was successfully demonstrated by Jung et al. in 2001 [1], who fused the Natronobacterium pharaonis NpSRII (Np seven-transmembrane retinylidene photoreceptor sensory rhodopsins II) and their cognate transducer HtrII to the cytoplasmic domain of the chemotaxis transducer EcTsr of Escherichia coli.
Rhodopsins are photoreactive, membrane-embedded proteins, which are found not only in archaea, but in eubacteria and microbes as well. In Natronobacterium pharaonis, the NpSRII contains a domain of seven membrane-spanning helices, which carry out two distinct functions: Firstly, they serve as photo-inducible ion-pumps and secondly, as actors in the chemotaxis signaling network [1].
Construct
We purchased the codon optimized version of the archeal photoreceptor NpSRII EcTsr-fusion from Geneart and cloned the sequence as biobrick into the standardized vector pSB1C3 ([http://partsregistry.org/Part:BBa_K422001 BBa_K422001]).
As membrane proteins are difficult to express at the appropriate concentrations, we decided to subclone the receptor into the IPTG-inducible plasmid pACT3 allowing us to adapt the expression level. The archeal receptor fusion was cloned into pACT3 via BamHI and HindIII. In addition to the BamHI site, the forward primer encodes for a ribosome binding site to ensure the optimal spacing to the start codon.
Forward (5'-3'): GTGGATCCAAGGAGATATACATATGGTTGGTCTGACCACCCTG
Reverse (5'-3'): GCAAGCTTTTAACCGCTATAAATTG
Chemotactic analysis
To observe chemotactic behaviour, cells were grown at 30 °C in Lysogeny Broth to on OD of 1.0. All-trans retinal has to be added to the media for NpSRII to change it's conformation into an active light absorbing pigment. Different levels of IPTG can influence the protein expression level.
Phototactic stimuli were delivered through a light pulse at 500 nm and cells tracked. You can find out more about microscopy and information processing here.
And don't forget, check out our results, the actual E. lemming.
Receptor analysis

To detect expression of the archeal light receptor, we carried out expression experiments with different IPTG inducer concentrations (0, 10, 100, 1'000 µM). We then loaded the crude cell extract on a 10 % SDS PAGE. Below you find the gel picture. At a fully induced condition, a band at approximately 68 KDa becomes more prominent. We assume this is the expressed archeal light receptor.
Due to the fact that this gel picture shows the whole protein fraction of an E. coli cell, we are planning to subclone ([http://partsregistry.org/Part:BBa_K422001 BBa_K422001]) into an expression vector containing a his-tag for protein purification.
An important experiment that we are planning to do before the Jamboree is to detect the insertion of the archeal light receptor into the membrane. For that, cells will have to be lysed after an induction phase and the membrane needs to be extracted by fractionated centrifugation. Using a monochromator, absorption of the membrane fraction at 500 nm (blue light) can be evaluated and compared between wild type and archeal light receptor expressing strain.
References
[1] [http://jb.asm.org/cgi/content/full/183/21/6365?maxtoshow=&hits=10&RESULTFORMAT=1&andorexacttitle=and&andorexacttitleabs=and&fulltext=An+archaeal+photosignal-transducing+module+mediates+phototaxis+in+%27%27Escherichia+&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&resourcetype=HWCIT: Jung, Spudich, Trivedi and Spudich: An archaeal photosignal-transducing module mediates phototaxis in Escherichia coli. Journal of bacteriology. 2001; 21].
 "
"