Team:Cambridge/Bioluminescence/Vibrio Characterisation
From 2010.igem.org
(→Arabinose to light) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Cambridge/Templates/headerMinimalprototype}} | {{:Team:Cambridge/Templates/headerMinimalprototype}} | ||
| - | {{:Team:Cambridge/Templates/headerbar|colour=# | + | {{:Team:Cambridge/Templates/headerbar|colour=#386abc|title=Project Vibrio: Characterisation}} |
This page describes characterisation for part [http://partsregistry.org/Part:BBa_K325909 BBa K325909], the [https://2010.igem.org/Team:Cambridge/Bioluminescence/Bacterial_Luciferases ''lux operon'' from ''Vibrio fischeri'']. | This page describes characterisation for part [http://partsregistry.org/Part:BBa_K325909 BBa K325909], the [https://2010.igem.org/Team:Cambridge/Bioluminescence/Bacterial_Luciferases ''lux operon'' from ''Vibrio fischeri'']. | ||
| Line 12: | Line 12: | ||
=Description= | =Description= | ||
{{:Team:Cambridge/Templates/RightImage|image=Cambridge-low.jpg|caption=E.Coli (Invitrogen TOP 10) cells transformed with [http://partsregistry.org/Part:BBa_K325909 BBa K325909] (blue light bulb) and [http://partsregistry.org/Part:BBa_K325219 BBa 325219] (red light bulb)}} | {{:Team:Cambridge/Templates/RightImage|image=Cambridge-low.jpg|caption=E.Coli (Invitrogen TOP 10) cells transformed with [http://partsregistry.org/Part:BBa_K325909 BBa K325909] (blue light bulb) and [http://partsregistry.org/Part:BBa_K325219 BBa 325219] (red light bulb)}} | ||
| - | This | + | This page described the lux operon from Vibrio fischeri. To relieve LuxR control we placed Lux C, D, A, B, E under the pBad promoter. |
| + | |||
| + | |||
| + | |||
| + | [[Image:250px-Cambridge-iGEMpixels.jpg|thumb|569px|center|'''Figure 1 - E.coli cells (Invitrogen TOP 10) transformed with [http://partsregistry.org/Part:BBa_K325909 BBa K325909] in a 96 well plate. ''']] | ||
=Arabinose to light= | =Arabinose to light= | ||
| Line 19: | Line 23: | ||
{{:Team:Cambridge/Templates/Nolineheader|header=Data}} | {{:Team:Cambridge/Templates/Nolineheader|header=Data}} | ||
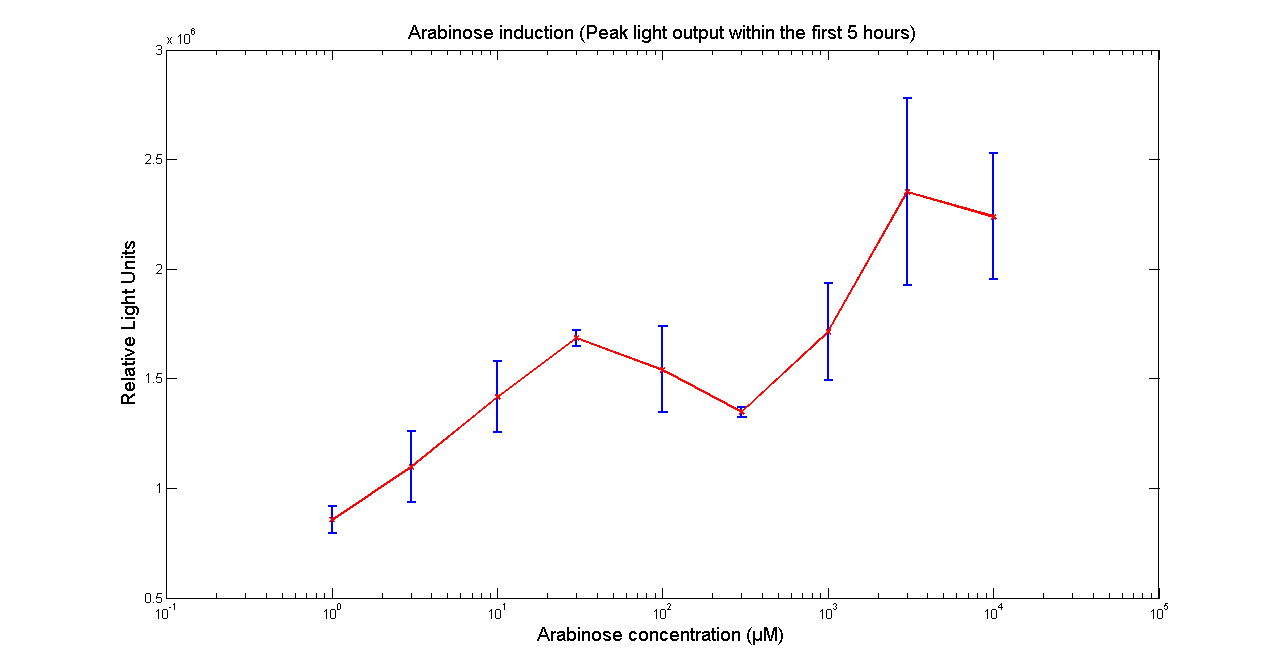
| - | [[Image:BBa_K325909Aracurve.png|thumb|569px|center|'''Figure 1 - Light output of <partinfo> | + | [[Image:BBa_K325909Aracurve.png|thumb|569px|center|'''Figure 1 - Light output of <partinfo>K325909</partinfo> as a function of Arabinose concentration in the media. The values correspond to the peak intensity within 5 hours of adding Arabinose to the media. Data points and error bars correspond to the mean and the standard deviation of 3 time repeats. ''']] |
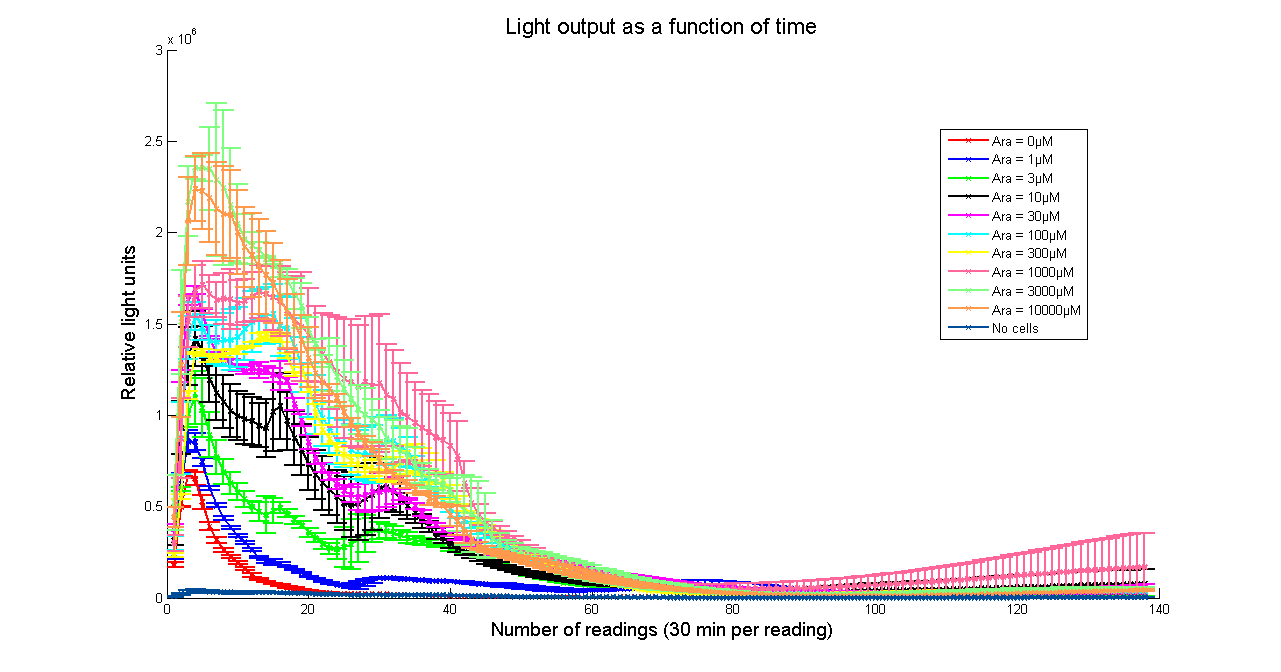
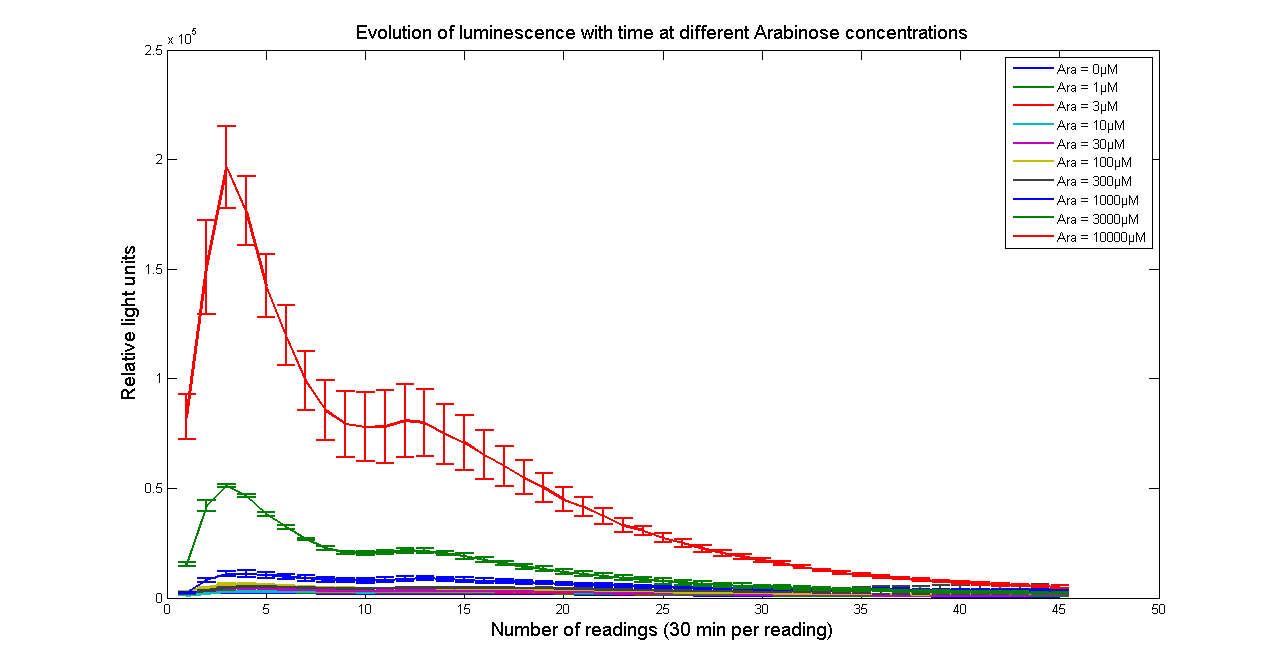
| - | [[Image:BBa_K325909timecourse.png|thumb|569px|center|'''Figure 2 - Evolution of luminescence with time at different Arabinose concentrations for <partinfo> | + | [[Image:BBa_K325909timecourse.png|thumb|569px|center|'''Figure 2 - Evolution of luminescence with time at different Arabinose concentrations for <partinfo>K325909</partinfo>. Measurements were taken every 30 minutes. Data points and error bars correspond to the mean and standard deviation of 3 time repeats. ''']] |
<center> | <center> | ||
{|{{Table}} | {|{{Table}} | ||
| Line 32: | Line 36: | ||
|} | |} | ||
</center> | </center> | ||
| - | |||
| - | |||
| - | |||
=H-NS mutants= | =H-NS mutants= | ||
| - | + | It has been shown that the expression of the Vibrio fischeri lux operon when cloned into E. coli was repressed. This repression was linked to the nucleoid protein H-NS. To investigate this effect we cloned the operon into mutant E.coli cells in which the expression of the H-NS protein had been modified. We used a [http://www.bmglabtech.com/products/microplate-reader/instruments.cfm?product_id=2 FLUOstar OPTIMA] microplate reader to quantify the light output. Protocols and plate reader settings used are given below. | |
{{:Team:Cambridge/Templates/Nolineheader|header=Data}} | {{:Team:Cambridge/Templates/Nolineheader|header=Data}} | ||
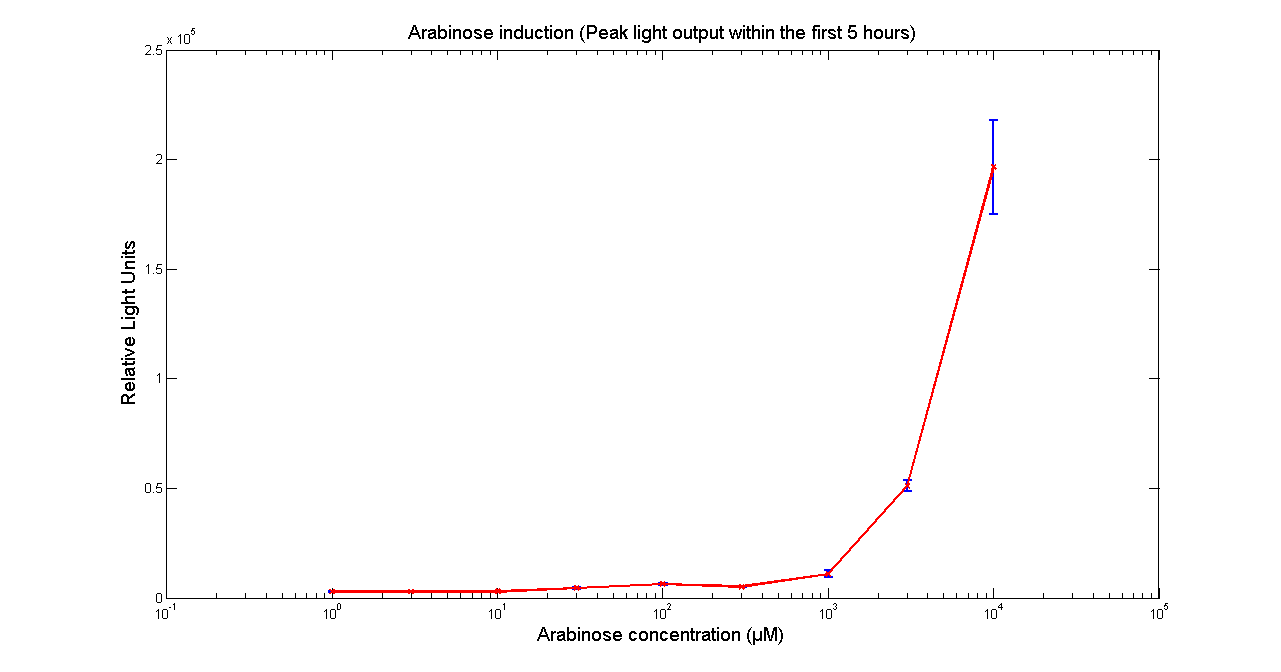
| - | ''' | + | [[Image:BBa_K325909AraK28.png|thumb|569px|center|'''Figure 1 - Peak light output from <partinfo>K325909</partinfo> cloned into H-NS mutant JM 230 H-NS -205::tn10. The data points and the error bar are the mean and standard deviation obtained by 3 tim repeats. ''']] |
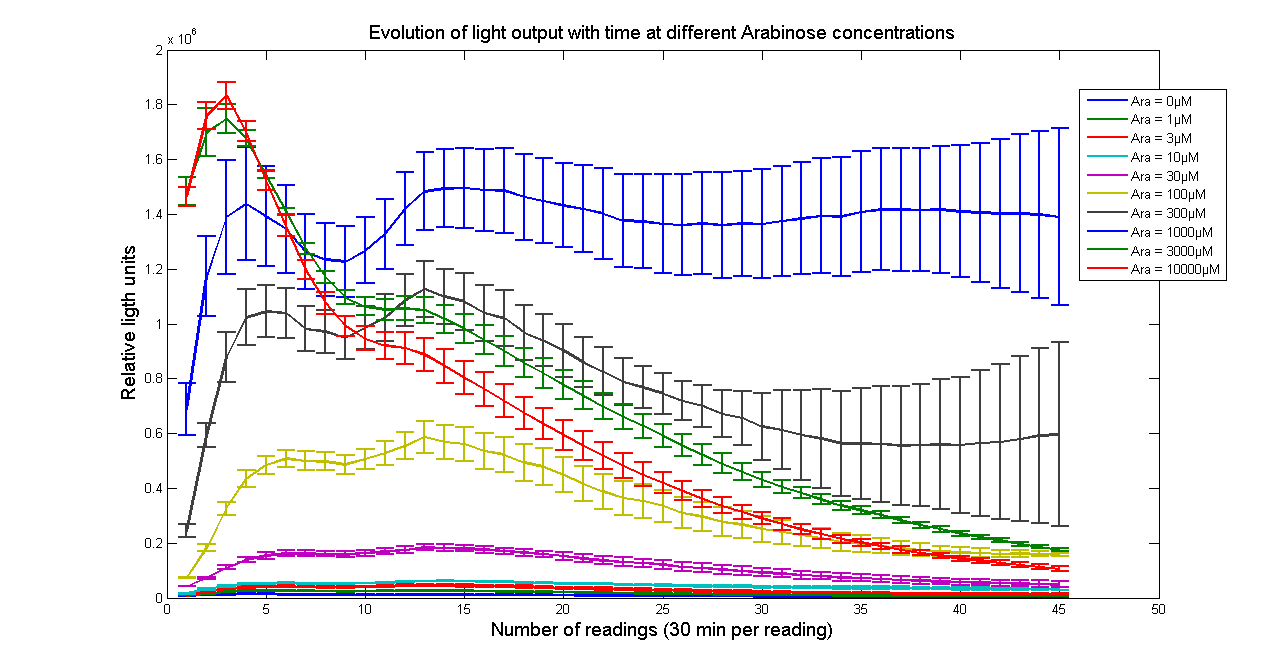
| - | + | [[Image:BBa_K325909timecourseK28.png|thumb|569px|center|'''Figure 2 - Evolution of light output from <partinfo>K325909</partinfo> cloned into H-NS mutant JM 230 H-NS -205::tn10 with time at different Arabinose concentrations. The data points and the error bar are the mean and standard deviation obtained by 3 time repeats. Measurements were taken every 30 minutes. ''']] | |
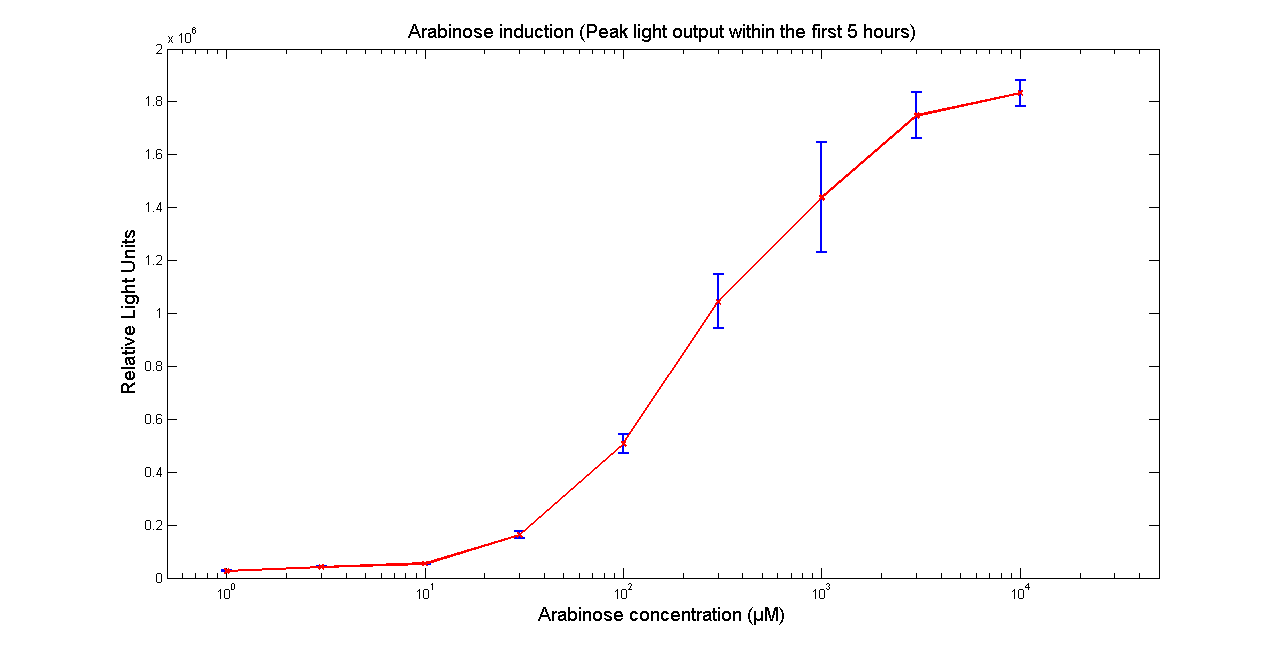
| - | + | [[Image:BBa_K325909AraR28.png|thumb|569px|center|'''Figure 3 - Figure 3 - Peak light output from <partinfo>K325909</partinfo> cloned into H-NS mutant [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan]. The data points and the error bar are the mean and standard deviation obtained by 3 tim repeats.''']] | |
| - | + | [[Image:BBa_K325909timecourseR28.png|thumb|569px|center|'''Figure 4 - Evolution of light output from <partinfo>K325909</partinfo> cloned into H-NS mutant [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan] with time at different Arabinose concentrations. The data points and the error bar are the mean and standard deviation obtained by 3 time repeats. Measurements were taken every 30 minutes.''']] | |
| - | The data points | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |- | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | '''Evolution of light output | + | |
| - | + | ||
| - | [http:// | + | |
| - | + | ||
| - | + | ||
<center> | <center> | ||
{|{{Table}} | {|{{Table}} | ||
| Line 102: | Line 53: | ||
!Date Uploaded | !Date Uploaded | ||
|- | |- | ||
| - | |[ | + | |[[Media:BBa_K325909Mutants.xls]] |
|Raw data from experiment | |Raw data from experiment | ||
|21/10/2010 | |21/10/2010 | ||
| Line 108: | Line 59: | ||
</center> | </center> | ||
| - | |||
| - | |||
=Compatibility= | =Compatibility= | ||
| - | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=cell ''Chassis:''] Device has been shown to work in ''Top 10 (Invitrogen)'' | + | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=cell ''Chassis:''] Device has been shown to work in ''Top 10 (Invitrogen)'', [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan] and H-NS mutant JM 230 H-NS -205::tn10.<br> |
[[Plasmid backbones|''Plasmids:'']] Device has been shown to work on ''<partinfo>pSB1C3</partinfo>'' <br> | [[Plasmid backbones|''Plasmids:'']] Device has been shown to work on ''<partinfo>pSB1C3</partinfo>'' <br> | ||
=References= | =References= | ||
| - | |||
| + | [http://www.jstor.org/stable/4449975 '''[1]:'''] J. Slock, (1995) Transformation Experiments Using Bioluminescence Genes of ''Vibrio fischeri'',''The American Biology Teacher'', '''57''', 225-227. | ||
| - | [http://www. | + | [http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.42.100188.001055 '''[2]:'''] E.A. Meighen (1988) Enzymes and genes from the ''lux'' operons of bioluminescent bacteria, ''Annual Reviews in Microbiology'' '''42''', 151-176. |
| + | [http://www.annualreviews.org/doi/pdf/10.1146/annurev.ge.28.120194.001001 '''[3]:'''] E.A. Meighen, (1994) Genetics of bacterial bioluminescence, ''Annual Reviews of Genetics'', '''28''', 117-139. | ||
| - | [http:// | + | [http://onlinelibrary.wiley.com/doi/10.1002/%28SICI%291099-1271%28199807/08%2913:4%3C185::AID-BIO486%3E3.0.CO;2-U/abstract '''[4]:'''] S. Ulitzur, (1998) H-NS controls the transcription of three promoters of ''Vibrio fischeri lux'' cloned in ''Escherichia coli'',''Journal of Bioluminescence and Chemiluminescence'', '''13'''(4), 185-188. |
| + | [http://www.nature.com/nature/journal/v444/n7117/full/nature05283.html '''[5]:'''] R.T. Dame ''et al.'', (2006) Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation,''Nature'', '''444''', 387-390. | ||
{{:Team:Cambridge/Templates/footer}} | {{:Team:Cambridge/Templates/footer}} | ||
Latest revision as of 23:24, 27 October 2010

This page describes characterisation for part [http://partsregistry.org/Part:BBa_K325909 BBa K325909], the lux operon from Vibrio fischeri.
Description

This page described the lux operon from Vibrio fischeri. To relieve LuxR control we placed Lux C, D, A, B, E under the pBad promoter.
Arabinose to light
This page describes the relationship between Arabinose concentration in the medium with light output. We used a [http://www.bmglabtech.com/products/microplate-reader/instruments.cfm?product_id=2 FLUOstar OPTIMA] microplate reader to quantify the light output. Protocol and plate reader settings used are given below.
Data
| Data | Notes | Date Uploaded |
|---|---|---|
| Media:BBa_K325909ArabinosetoLight.xls | Raw data from experiment | 21/10/2010 |
H-NS mutants
It has been shown that the expression of the Vibrio fischeri lux operon when cloned into E. coli was repressed. This repression was linked to the nucleoid protein H-NS. To investigate this effect we cloned the operon into mutant E.coli cells in which the expression of the H-NS protein had been modified. We used a [http://www.bmglabtech.com/products/microplate-reader/instruments.cfm?product_id=2 FLUOstar OPTIMA] microplate reader to quantify the light output. Protocols and plate reader settings used are given below.
Data
| Data | Notes | Date Uploaded |
|---|---|---|
| Media:BBa_K325909Mutants.xls | Raw data from experiment | 21/10/2010 |
Compatibility
[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=cell Chassis:] Device has been shown to work in Top 10 (Invitrogen), [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan] and H-NS mutant JM 230 H-NS -205::tn10.
Plasmids: Device has been shown to work on <partinfo>pSB1C3</partinfo>
References
[http://www.jstor.org/stable/4449975 [1]:] J. Slock, (1995) Transformation Experiments Using Bioluminescence Genes of Vibrio fischeri,The American Biology Teacher, 57, 225-227.
[http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.42.100188.001055 [2]:] E.A. Meighen (1988) Enzymes and genes from the lux operons of bioluminescent bacteria, Annual Reviews in Microbiology 42, 151-176.
[http://www.annualreviews.org/doi/pdf/10.1146/annurev.ge.28.120194.001001 [3]:] E.A. Meighen, (1994) Genetics of bacterial bioluminescence, Annual Reviews of Genetics, 28, 117-139.
[http://onlinelibrary.wiley.com/doi/10.1002/%28SICI%291099-1271%28199807/08%2913:4%3C185::AID-BIO486%3E3.0.CO;2-U/abstract [4]:] S. Ulitzur, (1998) H-NS controls the transcription of three promoters of Vibrio fischeri lux cloned in Escherichia coli,Journal of Bioluminescence and Chemiluminescence, 13(4), 185-188.
[http://www.nature.com/nature/journal/v444/n7117/full/nature05283.html [5]:] R.T. Dame et al., (2006) Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation,Nature, 444, 387-390.
 "
"