Team:USTC/Project/protein/bkgrd
From 2010.igem.org
Evelynzhang (Talk | contribs) |
|||
| (2 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | A fusion protein is the product of joining two genes or two proteins /peptides together. This is achieved through the creation of a fusion gene which is done through the new standard enzyme digestion of both the plasmids and the ligation of them. If it is two proteins that will be joined together, then a linker or spacer peptide will also be added due to our standard. This would usually make it more likely for the proteins to fold independently and behave as it should be. | + | __NOTOC__ |
| + | {{Template:USTCiGEM2010_header}} | ||
| + | |||
| + | <html> | ||
| + | <style> | ||
| + | #transparent{ | ||
| + | position:relative; | ||
| + | left:255px; | ||
| + | width:675px; | ||
| + | height:2000px; | ||
| + | padding:10px; | ||
| + | background-image:url(https://static.igem.org/mediawiki/2010/a/a0/Transppa.png); | ||
| + | background-repeat:repeat; | ||
| + | filter:alpha(opacity=90); | ||
| + | opacity:0.90; | ||
| + | border-bottom-right-radius:20px; | ||
| + | border-bottom-left-radius:20px; | ||
| + | border-top-left-radius:20px; | ||
| + | border-top-right-radius:20px; | ||
| + | } | ||
| + | </style> | ||
| + | <div id="transparent"> | ||
| + | </html> | ||
| + | <html> | ||
| + | <style> | ||
| + | .firstletter{ | ||
| + | font-size:3em; | ||
| + | line-height:0.95; | ||
| + | float:left; | ||
| + | </style> | ||
| + | </html> | ||
| + | <p ><span class="firstletter">A</span> | ||
| + | fusion protein is the product of joining two genes or two proteins /peptides together. This is achieved through the creation of a fusion gene which is done through the new standard enzyme digestion of both the plasmids and the ligation of them. If it is two proteins that will be joined together, then a linker or spacer peptide will also be added due to our standard. This would usually make it more likely for the proteins to fold independently and behave as it should be.</P> | ||
According to the standard assembly, biobrick is flanked by restriction sites, comprised of the BioBrick Prefix and Suffix, between the EcoRI and XbaI cutting sites on the left and SpeI and PstI on the right. However, in our experiments, the biobricks were produced by the new standard, containing the SacI, SapI and EarI cutting sites. Though the standard assembly is designed reasonable, it sticks into troubles when coming to the fusion proteins. With the help of our new standard, fusion proteins, which are used for the identification, localization and purification of BMC, were better expressed and utilized. Our project consist of a RFP protein, a GFP protein, a GST protein or HIS protein. | According to the standard assembly, biobrick is flanked by restriction sites, comprised of the BioBrick Prefix and Suffix, between the EcoRI and XbaI cutting sites on the left and SpeI and PstI on the right. However, in our experiments, the biobricks were produced by the new standard, containing the SacI, SapI and EarI cutting sites. Though the standard assembly is designed reasonable, it sticks into troubles when coming to the fusion proteins. With the help of our new standard, fusion proteins, which are used for the identification, localization and purification of BMC, were better expressed and utilized. Our project consist of a RFP protein, a GFP protein, a GST protein or HIS protein. | ||
| Line 11: | Line 43: | ||
[[Image:USTC2010_PduP-GFP.JPG]] | [[Image:USTC2010_PduP-GFP.JPG]] | ||
| - | Fig 2a:Localization of fusion proteins by fluorescence microscopy of cells producing eGFP (A) and PduP[1-18]eGFP (B). Arrows point to areas of localized brighter green fluorescence[http://www.pnas.org/content/107/16/7509.long | + | Fig 2a:Localization of fusion proteins by fluorescence microscopy of cells producing eGFP (A) and PduP[1-18]eGFP (B). Arrows point to areas of localized brighter green fluorescence[http://www.pnas.org/content/107/16/7509.long] |
Third, a similar set of studies was done by fusing N-terminal sections of PduV to GFP(Fig 3a). As it is reported, PduV localized to cup-like structures upon the outer surface of the BMCs. 98 amino acids from the N terminus of PduV were fused to GFP(Fig 3b. PduV (green) and PduA-B-J-K-N-U structures (red) are seen to interact dynamically and move in a coordinated manner within elongating cells. Time between frames, 1 min). In addition, fusion proteins PduV-GST and PduV-HIS were expected to purify the BMC protein because of the affinity to special elution buffer. | Third, a similar set of studies was done by fusing N-terminal sections of PduV to GFP(Fig 3a). As it is reported, PduV localized to cup-like structures upon the outer surface of the BMCs. 98 amino acids from the N terminus of PduV were fused to GFP(Fig 3b. PduV (green) and PduA-B-J-K-N-U structures (red) are seen to interact dynamically and move in a coordinated manner within elongating cells. Time between frames, 1 min). In addition, fusion proteins PduV-GST and PduV-HIS were expected to purify the BMC protein because of the affinity to special elution buffer. | ||
Latest revision as of 22:18, 27 October 2010
A fusion protein is the product of joining two genes or two proteins /peptides together. This is achieved through the creation of a fusion gene which is done through the new standard enzyme digestion of both the plasmids and the ligation of them. If it is two proteins that will be joined together, then a linker or spacer peptide will also be added due to our standard. This would usually make it more likely for the proteins to fold independently and behave as it should be.
According to the standard assembly, biobrick is flanked by restriction sites, comprised of the BioBrick Prefix and Suffix, between the EcoRI and XbaI cutting sites on the left and SpeI and PstI on the right. However, in our experiments, the biobricks were produced by the new standard, containing the SacI, SapI and EarI cutting sites. Though the standard assembly is designed reasonable, it sticks into troubles when coming to the fusion proteins. With the help of our new standard, fusion proteins, which are used for the identification, localization and purification of BMC, were better expressed and utilized. Our project consist of a RFP protein, a GFP protein, a GST protein or HIS protein.
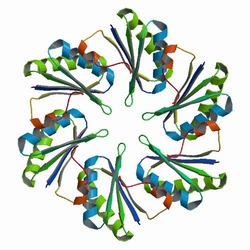
First, the localization of targeted proteins to the shell protein assemblies was investigated by the fusion of RFP-PduA. The structure of PduA can be seen in Fig 1a. Through the expression of PduA, we have known the structure of BMC(Fig 1b). We cloned the RFP gene to the 5’ end of pduA. When the fusion protein was produced, patches of red were observed throughout the cell.
Fig 1a: The Biological Assembly Image of PduA[http://www.rcsb.org/pdb/explore/explore.do?structureId=3NGK]
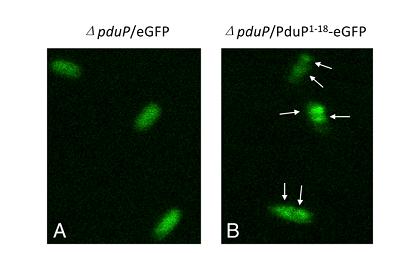
Second, we know that a N-terminal region of PduP is required for efficient packaging into the BMC. To test this, fusion proteins (P[1-14]-GFP, P[1-18]-GFP, P[1-64]-GFP) were used. (Fig 2a).Although P[1-14] and P[1-18] were also be taken into consideration, we created P[1-64]-GFP fusion protein in the end to study the transportation of BMC and examined subcellular localization of it by fluorescence and electron microscopy.
Fig 2a:Localization of fusion proteins by fluorescence microscopy of cells producing eGFP (A) and PduP[1-18]eGFP (B). Arrows point to areas of localized brighter green fluorescence[http://www.pnas.org/content/107/16/7509.long]
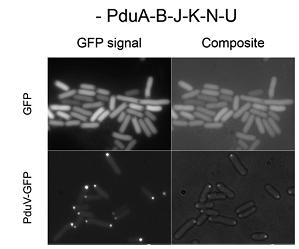
Third, a similar set of studies was done by fusing N-terminal sections of PduV to GFP(Fig 3a). As it is reported, PduV localized to cup-like structures upon the outer surface of the BMCs. 98 amino acids from the N terminus of PduV were fused to GFP(Fig 3b. PduV (green) and PduA-B-J-K-N-U structures (red) are seen to interact dynamically and move in a coordinated manner within elongating cells. Time between frames, 1 min). In addition, fusion proteins PduV-GST and PduV-HIS were expected to purify the BMC protein because of the affinity to special elution buffer.
Fig 3a:Cellular Distribution of PduV Is Altered by the Presence of PduA-B-J-K-N-U Structures[http://www.ncbi.nlm.nih.gov/pubmed/20417607]
 "
"