Team:UT-Tokyo/Sudoku experiments
From 2010.igem.org
(→Parts list) |
|||
| (29 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
= '''Sudoku''' = | = '''Sudoku''' = | ||
| - | + | <html> | |
| - | + | <div> | |
| - | + | <ul id="inpagemenu"> | |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_abstract" id="abstract">Introduction</a></li> | |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_construct" id="construct">System</a></li> | |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_modeling" id="modeling">Modeling</a></li> | |
| - | + | <li><span>Experiments</span></li> | |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_perspective" id="perspective">Perspective</a></li> | |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_reference" id="reference">Reference</a></li> | |
| + | </ul> | ||
| + | </div> | ||
| + | <div id="clear"></div> | ||
| + | </html> | ||
| + | =='''Experiments'''== | ||
=== '''Assay''' === | === '''Assay''' === | ||
| - | 1. [[#Terminator leak]] | + | 1. [[#Terminator leak|Terminator leak]] |
| - | 2. [[#Phage MS2]] | + | 2. [[#Phage MS2|Phage MS2]] |
| - | 3. [[#Location sequence]] | + | 3. [[#Location sequence|Location sequence]] |
==='''Parts Making'''=== | ==='''Parts Making'''=== | ||
| - | 1. [[#Terminator leak(Parts)]] | + | 1. [[#Terminator leak(Parts Making)|Terminator leak(Parts Making)]] |
| - | 2. [[#Phage MS2(Parts)]] | + | 2. [[#Phage MS2(Parts Making)|Phage MS2(Parts Making)]] |
| - | 3. [[#Location sequence(Parts)]] | + | 3. [[#Location sequence(Parts Making)|Location sequence(Parts Making)]] |
| - | 4. [[#Hin]] | + | 4. [[#Hin|Hin]] |
| - | 5. [[#flpe]] | + | 5. [[#flpe|flpe]] |
| - | |||
==='''Detail Note Book'''=== | ==='''Detail Note Book'''=== | ||
| + | |||
| + | |||
| + | |||
= Assay = | = Assay = | ||
== [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_LeakSw Terminator leak] == | == [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_LeakSw Terminator leak] == | ||
| - | [[Image: | + | [[Image:Sudoku_Assay_terminator_4.png|200px|thumb|Terminator leak switch assay]] |
| + | |||
| + | |||
| + | To construct 4C3 leak-switch, which is indispensable to determine the numbers when solving Sudoku, the following system is required: do not express enough amount of Cre gene that has two or more terminators on the upstream, and express enough amount of Cre gene that has only one terminator there. For this purpose, it is neccessary to use appropriate terminator. | ||
| + | |||
| + | The aim of terminator leakiness assay is to make sure whether our terminator is appropriate or not. We measured the fluorescence of GFP expressed when Cre protein recombine DNA construct. When the fluorescence of expressed GFP depends on the existence of Cre gene in this construct, our system works properly. However, in the first trial, the fluorescence of GFP cannot be observed in our construct. We considered whether lox sequence, cre coding sequence or both of them have some errors, and performed some experiments to make sure our hypothesis. As a result, it elucidated that lox sequence is not functional. | ||
== [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_MS2 Phage MS2] == | == [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_MS2 Phage MS2] == | ||
| - | [[Image: | + | [[Image:Sudoku_Assay_MS2_6.png|200px|thumb|Phage MS2 expression check assay]] |
We use MS2 phage to transmit information of location and number. | We use MS2 phage to transmit information of location and number. | ||
| Line 63: | Line 75: | ||
== [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_LocSq Location sequence] == | == [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_LocSq Location sequence] == | ||
| + | [[Image:Sudoku_Assay_location_6.png|200px|thumb|Translational repression assay: antisense RNA]] | ||
| + | |||
The object of the assay is to test whether antisense RNA used in our construct works or not. In order to block the unnecessary information transformed by the virus from the other grid, we use antisense RNA to block ribosome to bind the region around the ribosome binding site (rbs) and prevent the expression of protein. In our construct, information is carried by virus and antisense RNA is transcribed constantly inside the cell. Once the unnecessary information was transformed, the antisense will come and shut out all the RNA chain excluded by virus. | The object of the assay is to test whether antisense RNA used in our construct works or not. In order to block the unnecessary information transformed by the virus from the other grid, we use antisense RNA to block ribosome to bind the region around the ribosome binding site (rbs) and prevent the expression of protein. In our construct, information is carried by virus and antisense RNA is transcribed constantly inside the cell. Once the unnecessary information was transformed, the antisense will come and shut out all the RNA chain excluded by virus. | ||
In this assay, we used pBAD as the promoter to start translating grid information which will be transformed by virus in our construct. The strength of the promoter depends on the concentration of arabinose. On the other hand, we used c-pro as the promoter to start translating antisense RNA. This c-pro is the strongest constitutive promoter submitted in igem parts. In assay I, we examined the relative strength of pBAD (with eight different concentration of arabinose) and c-pro. By using the proper concentration of arabinose which was determined by assay I, in assay II, we inspect observe whether our antisense RNA works or not. In these two assay, gfp was used as a reporter protein. | In this assay, we used pBAD as the promoter to start translating grid information which will be transformed by virus in our construct. The strength of the promoter depends on the concentration of arabinose. On the other hand, we used c-pro as the promoter to start translating antisense RNA. This c-pro is the strongest constitutive promoter submitted in igem parts. In assay I, we examined the relative strength of pBAD (with eight different concentration of arabinose) and c-pro. By using the proper concentration of arabinose which was determined by assay I, in assay II, we inspect observe whether our antisense RNA works or not. In these two assay, gfp was used as a reporter protein. | ||
= Parts Making = | = Parts Making = | ||
| - | == Terminator leak(Parts) == | + | == Terminator leak(Parts Making) == |
| - | == Phage MS2(Parts) == | + | [[Image:Sudoku_Assay_terminator_3.png|200px|thumb|Terminator leak switch assay]] |
| + | To realize 4C3 leak switch, we should choose proper terminator which terminates transcription when connected two or | ||
| + | |||
| + | more but leak when single. | ||
| + | |||
| + | We made "A-M-Ro/Char" assay (see Fig, named from the famous animation, "MOBILE SUIT GUNDAM") to select proper | ||
| + | |||
| + | terminator: | ||
| + | |||
| + | '''Aoi''' | ||
| + | |||
| + | no terminator | ||
| + | |||
| + | -> cre protein express rapidly | ||
| + | |||
| + | -> lox site is removed rapidly | ||
| + | |||
| + | -> gfp may expression rapidly | ||
| + | |||
| + | |||
| + | '''Murasaki''' | ||
| + | |||
| + | one terminator | ||
| + | |||
| + | -> cre protein express slowly | ||
| + | |||
| + | -> lox site is removed slowly | ||
| + | |||
| + | -> gfp may expression slowly | ||
| + | |||
| + | |||
| + | '''Rokujyo''' | ||
| + | |||
| + | two terminator | ||
| + | |||
| + | -> cre protein can't express | ||
| + | |||
| + | -> lox site remain | ||
| + | |||
| + | -> gfp expression may not express | ||
| + | |||
| + | |||
| + | First we use single terminator BBa_B1006. | ||
| + | Then other terminators are tested: 80%-terminate terminator and 99%-terminate terminator to determine the best | ||
| + | |||
| + | threshold to realize terminator leak switch. | ||
| + | |||
| + | == Phage MS2(Parts Making) == | ||
[[Image:Sudoku_MS2-make_2.jpg|200px|thumb|How to make MS2 parts]] | [[Image:Sudoku_MS2-make_2.jpg|200px|thumb|How to make MS2 parts]] | ||
| Line 98: | Line 159: | ||
| - | == Location sequence(Parts) == | + | == Location sequence(Parts Making) == |
| + | ===Parts making=== | ||
| + | [[Image:Sudoku_location_making.png|200px|thumb|How to make location parts]] | ||
| + | |||
| + | Ligate small parts - recognation site, location site and rbs. | ||
| + | |||
| + | ===Expression check=== | ||
| + | [[Image:Sudoku_Assay_location_4.png|200px|thumb|Location sequence]] | ||
| + | |||
| + | This assay testifies whether translation repression by location sequence go well. | ||
| + | |||
| + | First we check the ability of pBAD, which is induced by arabinose, by the expression of tetracycline-tolerance | ||
| + | |||
| + | protein. | ||
| + | |||
| + | Then to check translation repression, we make assay which express gfp only when translation repression can’t be | ||
| + | |||
| + | occurring. | ||
== Hin == | == Hin == | ||
| Line 152: | Line 230: | ||
| - | |||
| - | |||
| - | |||
= Parts list = | = Parts list = | ||
| Line 186: | Line 261: | ||
|- | |- | ||
| 7 || rbs (rev) || | | 7 || rbs (rev) || | ||
| - | [http://partsregistry.org/wiki/index.php?title=Part: | + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_J44001 BBa_J44001] |
|| plate1-1J || pSB1A2 || 15bp | || plate1-1J || pSB1A2 || 15bp | ||
|- | |- | ||
| Line 261: | Line 336: | ||
|| --- || --- || about 1.2kbp | || --- || --- || about 1.2kbp | ||
|} | |} | ||
| + | |||
| + | |||
| + | <br><br> | ||
| + | |||
| + | <h2> | ||
| + | [http://igem-ut.net/2010/now/ Latest Experiments] | ||
| + | </h2> | ||
| + | <p> | ||
| + | Our Latest Experiment & results. Available from [http://igem-ut.net/2010/now/ here]. | ||
| + | </p> | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | = Detail Note Book = | ||
| + | |||
| + | |||
| + | Detail protocols about Sudoku project are: | ||
| + | |||
| + | [https://2010.igem.org/Team:UT-Tokyo/Sudoku_notebook_6 June]. | ||
| + | |||
| + | [https://2010.igem.org/Team:UT-Tokyo/Sudoku_notebook July]. | ||
| + | |||
| + | [https://2010.igem.org/Team:UT-Tokyo/Sudoku_notebook_2 August]. | ||
| + | |||
| + | [https://2010.igem.org/Team:UT-Tokyo/Sudoku_notebook_8 September - October]. | ||
| + | |||
| + | |||
{{UT-Tokyo_Foot}} | {{UT-Tokyo_Foot}} | ||
Latest revision as of 10:46, 13 February 2011


Sudoku
Experiments
Assay
2. Phage MS2
Parts Making
1. Terminator leak(Parts Making)
3. Location sequence(Parts Making)
4. Hin
5. flpe
Detail Note Book
Assay
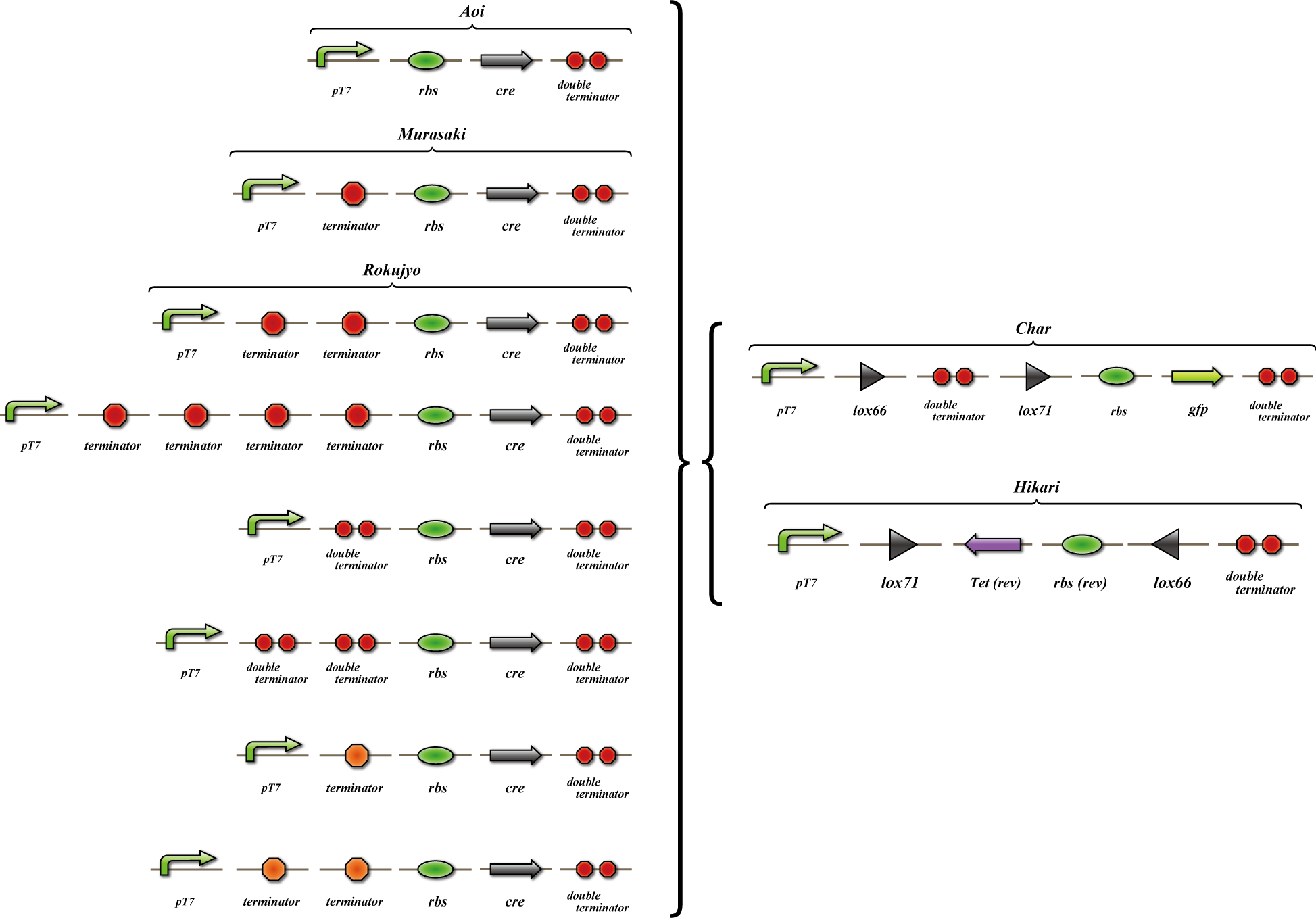
Terminator leak
To construct 4C3 leak-switch, which is indispensable to determine the numbers when solving Sudoku, the following system is required: do not express enough amount of Cre gene that has two or more terminators on the upstream, and express enough amount of Cre gene that has only one terminator there. For this purpose, it is neccessary to use appropriate terminator.
The aim of terminator leakiness assay is to make sure whether our terminator is appropriate or not. We measured the fluorescence of GFP expressed when Cre protein recombine DNA construct. When the fluorescence of expressed GFP depends on the existence of Cre gene in this construct, our system works properly. However, in the first trial, the fluorescence of GFP cannot be observed in our construct. We considered whether lox sequence, cre coding sequence or both of them have some errors, and performed some experiments to make sure our hypothesis. As a result, it elucidated that lox sequence is not functional.
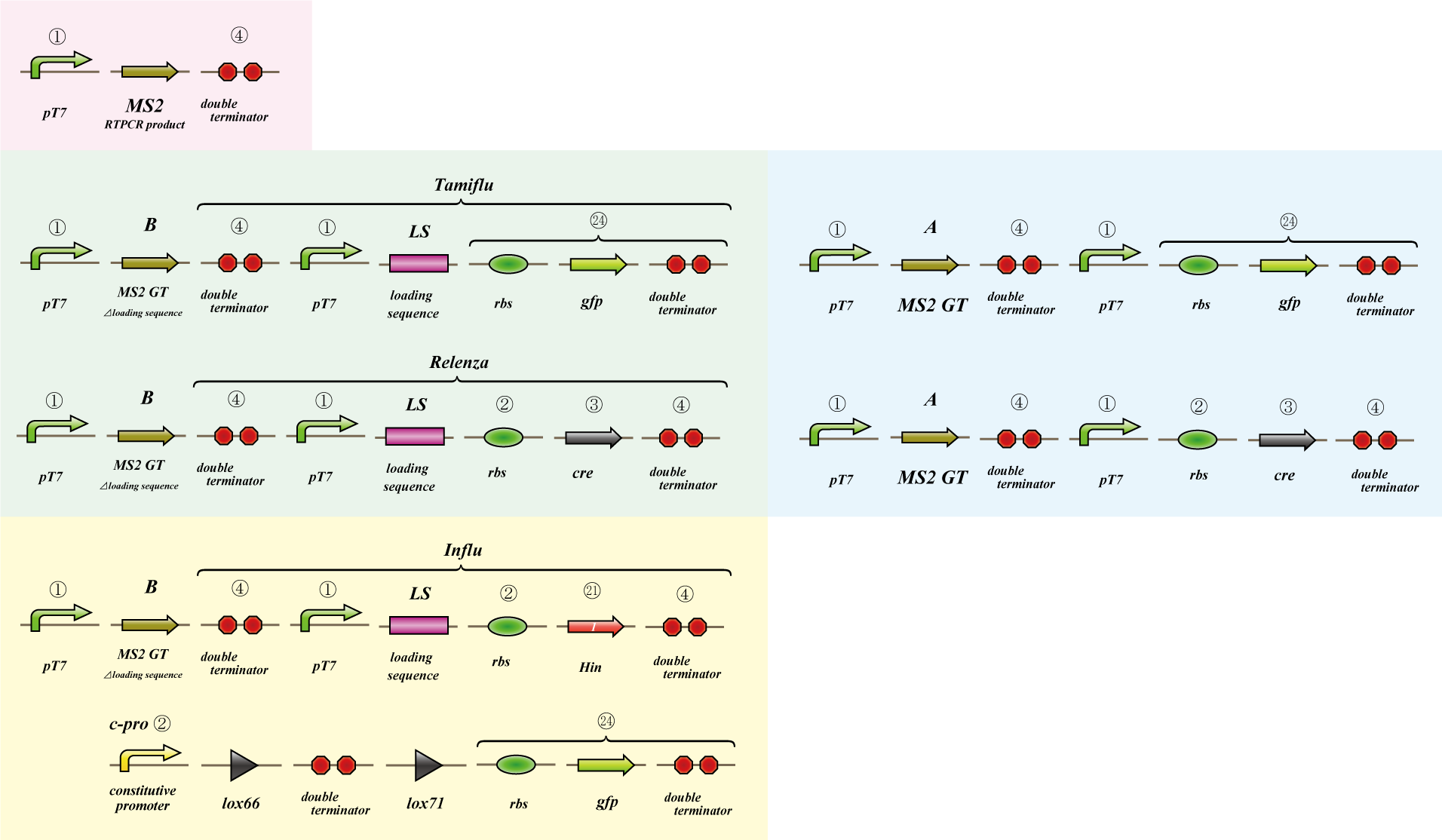
Phage MS2
We use MS2 phage to transmit information of location and number. The expression of the phage should start after 4C3 leak switch turns on. MS2 phage transport RNA which has loding sequence, so we knock out self assembly and made our E.coli translate loading seaquence at another point. To check whether transportion go correctly, we made assay.
i. Phage expression assay (red region)
Check whether RT-PCR product can be translated into MS2 phage.
ii. Packaging assay (blue & green region)
MS2 phage package RNA which has loading sequence. Using this character, we check whether RNA which has loading sequence and other coding region (gfp, cre) can be packaged correctly.
iii. Infection assay (yellow & green region)
We make the assay, which express gfp only when cre protein is expressed correctly. By using this part, we check whether E.coli can be infected with the MS2 phage made in assay ii as the fluorescence of gfp.
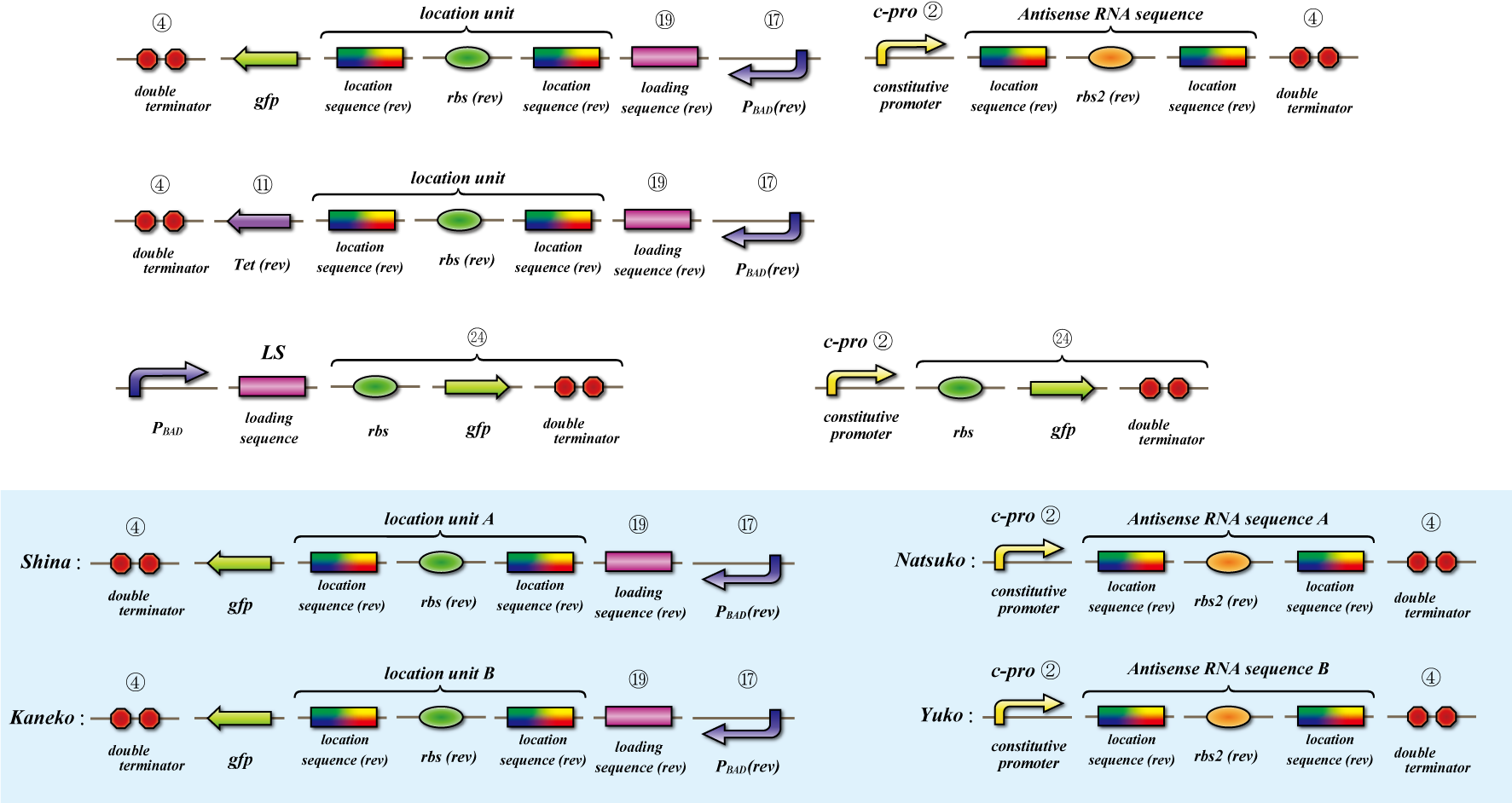
Location sequence
The object of the assay is to test whether antisense RNA used in our construct works or not. In order to block the unnecessary information transformed by the virus from the other grid, we use antisense RNA to block ribosome to bind the region around the ribosome binding site (rbs) and prevent the expression of protein. In our construct, information is carried by virus and antisense RNA is transcribed constantly inside the cell. Once the unnecessary information was transformed, the antisense will come and shut out all the RNA chain excluded by virus. In this assay, we used pBAD as the promoter to start translating grid information which will be transformed by virus in our construct. The strength of the promoter depends on the concentration of arabinose. On the other hand, we used c-pro as the promoter to start translating antisense RNA. This c-pro is the strongest constitutive promoter submitted in igem parts. In assay I, we examined the relative strength of pBAD (with eight different concentration of arabinose) and c-pro. By using the proper concentration of arabinose which was determined by assay I, in assay II, we inspect observe whether our antisense RNA works or not. In these two assay, gfp was used as a reporter protein.
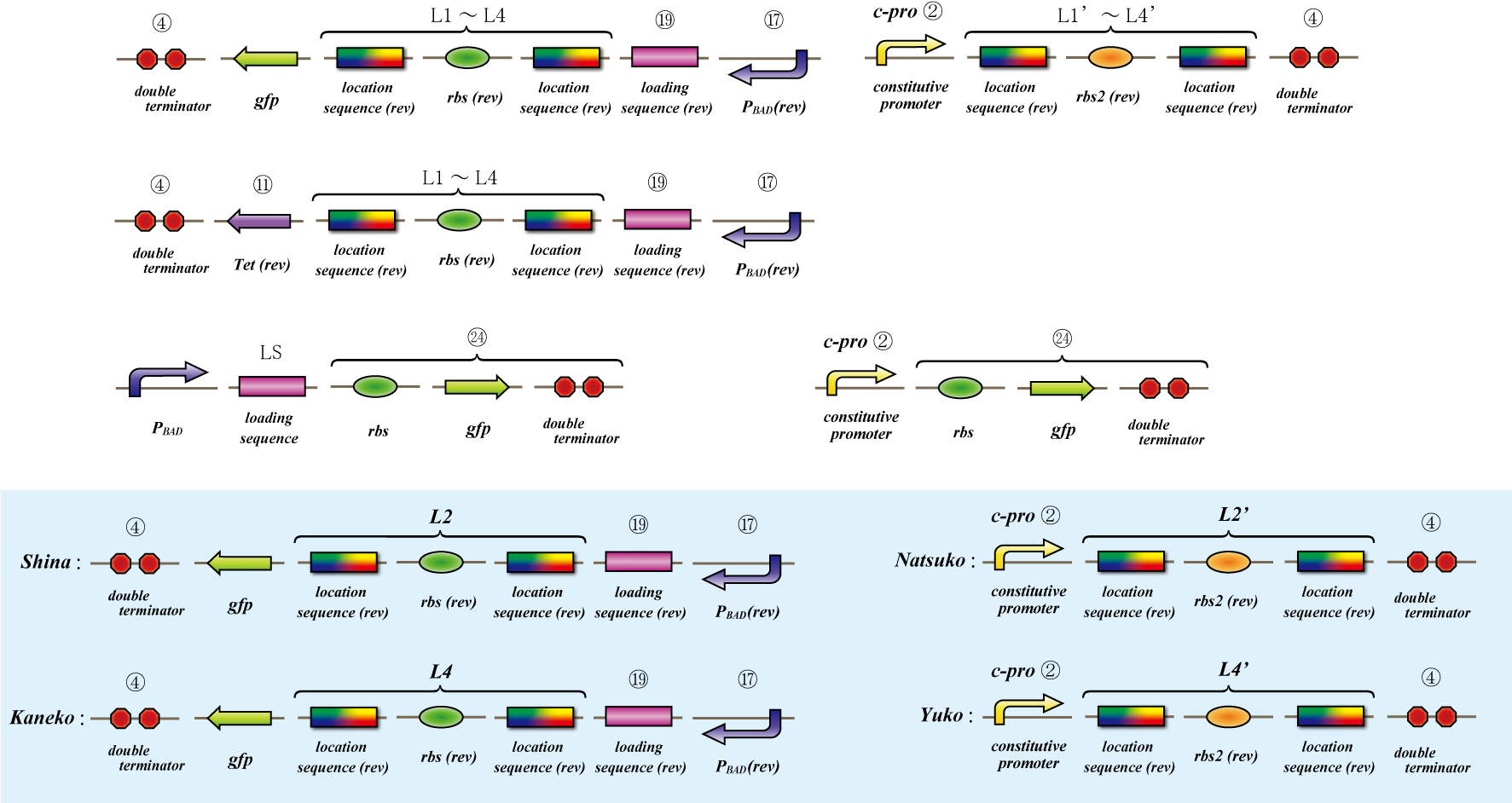
Parts Making
Terminator leak(Parts Making)
To realize 4C3 leak switch, we should choose proper terminator which terminates transcription when connected two or
more but leak when single.
We made "A-M-Ro/Char" assay (see Fig, named from the famous animation, "MOBILE SUIT GUNDAM") to select proper
terminator:
Aoi
no terminator
-> cre protein express rapidly
-> lox site is removed rapidly
-> gfp may expression rapidly
Murasaki
one terminator
-> cre protein express slowly
-> lox site is removed slowly
-> gfp may expression slowly
Rokujyo
two terminator
-> cre protein can't express
-> lox site remain
-> gfp expression may not express
First we use single terminator BBa_B1006.
Then other terminators are tested: 80%-terminate terminator and 99%-terminate terminator to determine the best
threshold to realize terminator leak switch.
Phage MS2(Parts Making)
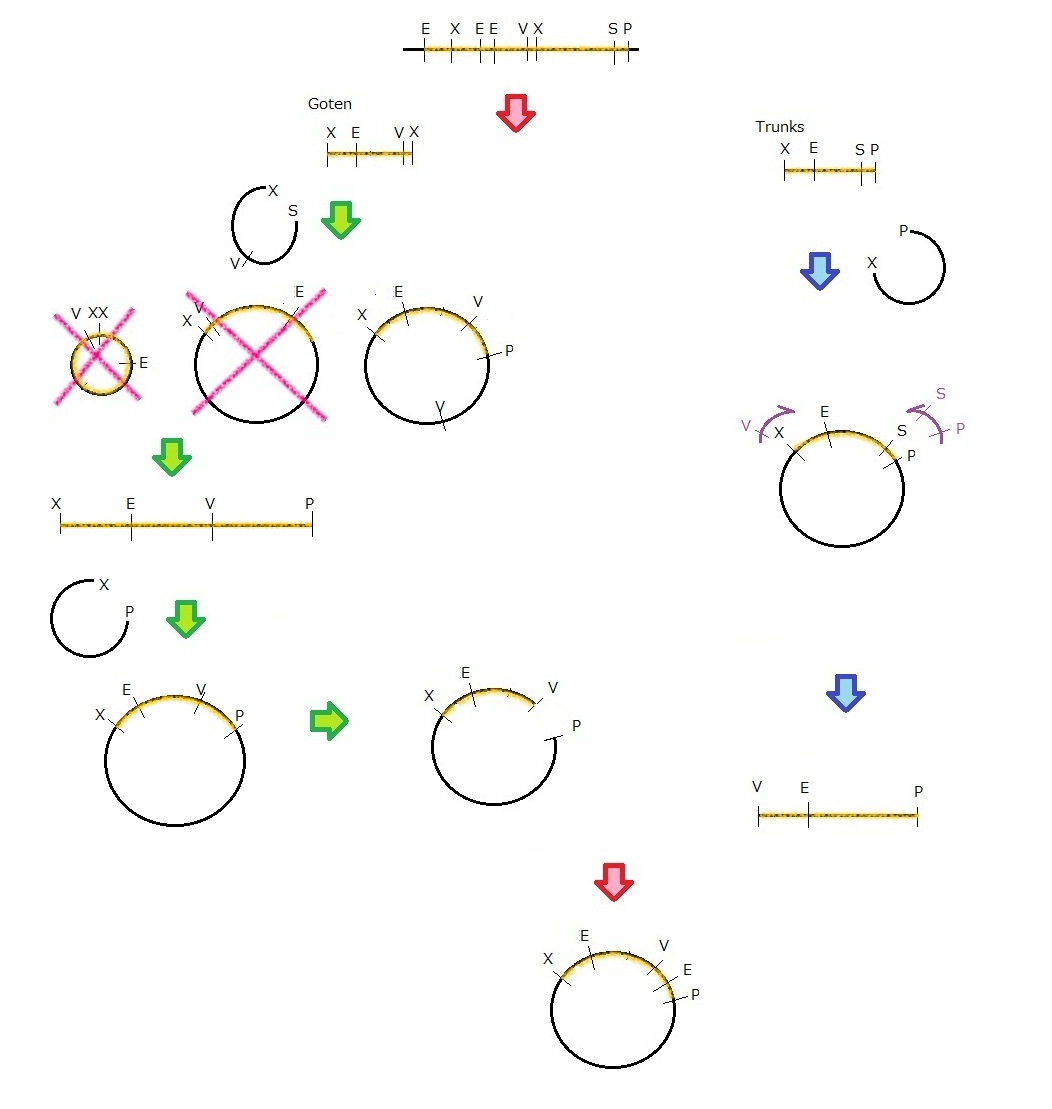
We get MS2 gene RT-PCR product. This original product include a lot of restricted enzyme site: two EcoRI site, two XbaI site and one VspI site.
To run our project, we don’t have to remove EcoRI site in the region. So we modified XbaI site by PCR as the following method:
1. We divided RT-PCR product into two parts by XP enzyme digestion:
E-X-E-E-V-X -> “Goten”
X-S-P -> “Tranks”
(named from the famous animation, "DRAGON BALL")
2. Insert is ligated with the vector, chloramphenicol-tolerance:
Goten -> EX vector
Tranks -> XP vector
3. Tranks -> PCR adding V-X region
4. Each part -> VP enzyme digestion, ligation each other
Location sequence(Parts Making)
Parts making
Ligate small parts - recognation site, location site and rbs.
Expression check
This assay testifies whether translation repression by location sequence go well.
First we check the ability of pBAD, which is induced by arabinose, by the expression of tetracycline-tolerance
protein.
Then to check translation repression, we make assay which express gfp only when translation repression can’t be
occurring.
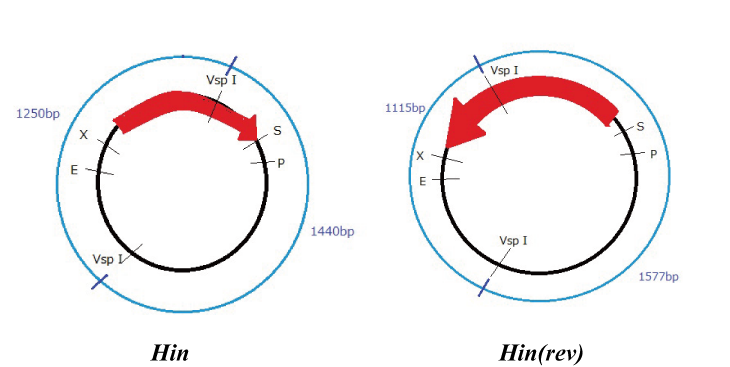
Hin
Parts making (reverse)
We get Hin parts from HQ (BBa_J31000).
We use this part as the form of reverse, so we made this part reverse by PCR.
To check PCR is done correctly, we did VspI digestion.
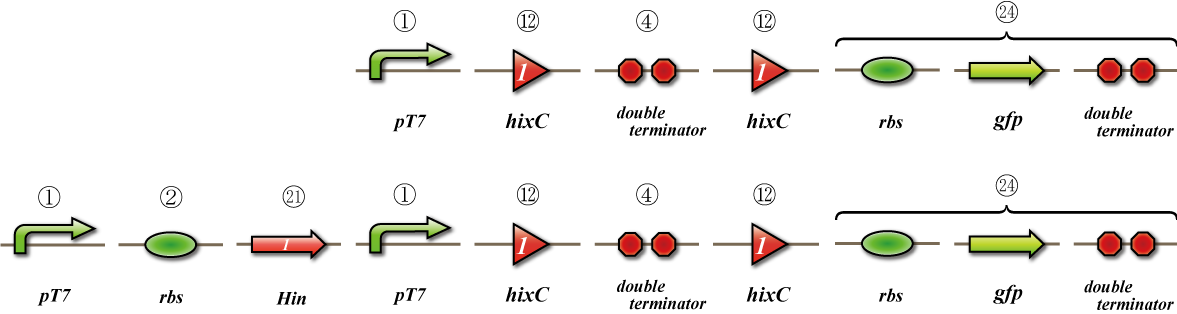
Expression check
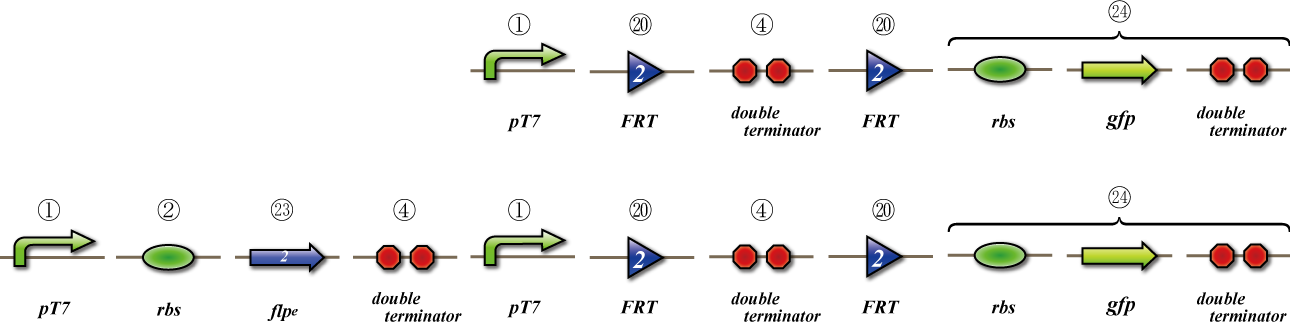
To check whether flpe works correctly, we made assay shown in Fig, similar to the assay of flpe check.
The top construct is a nagative control. GFP can't be expressed because of the double terminators.
The bottom construct express GFP when Hin works correctly. When Hin is expressed correctly, Hin recognaize the hix site and double terminator which terminates the expression of gfp is removed, so GFP may be expressed.
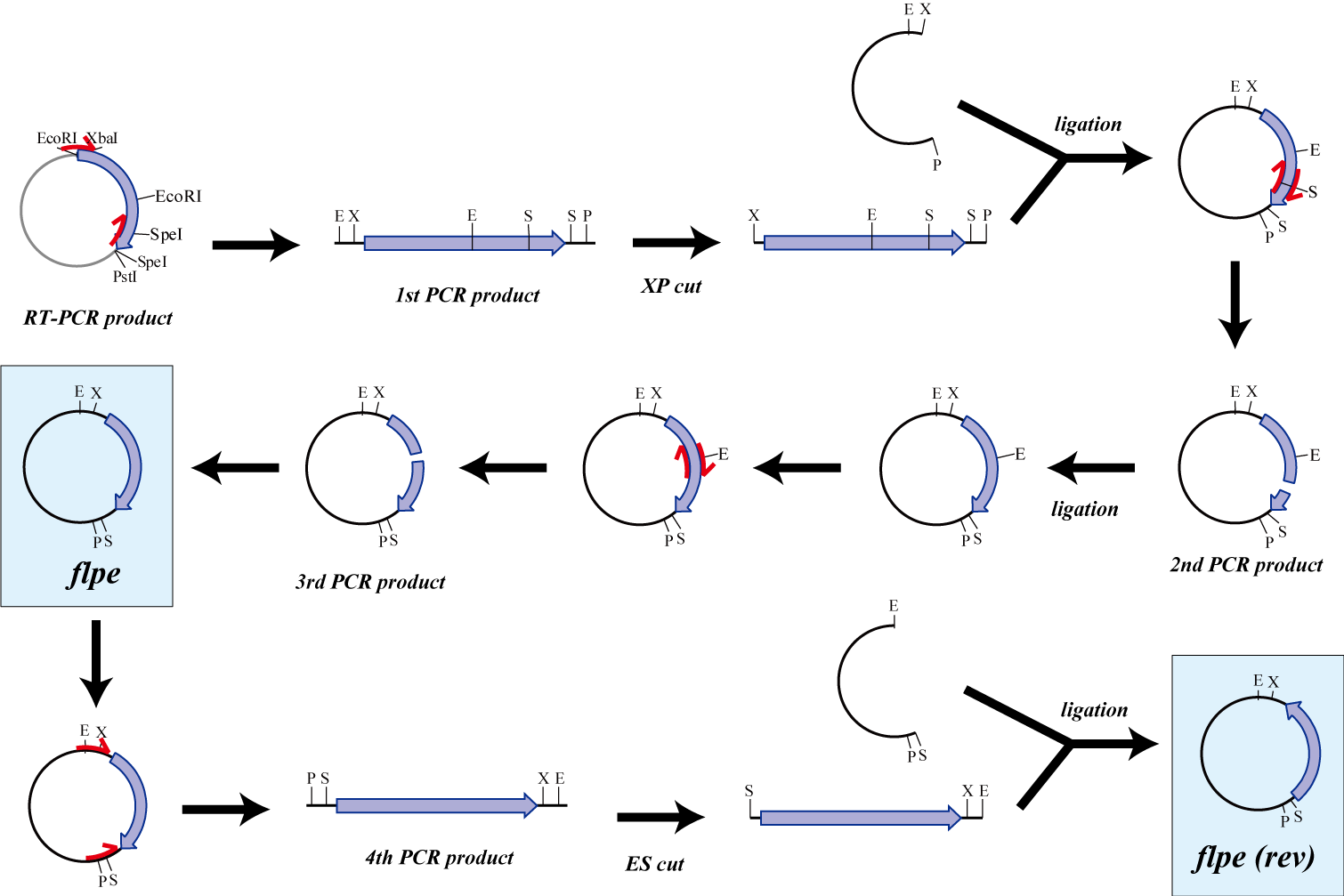
flpe
The original flpe include two restriction enzyme sites(EcoRI, SpeI).
We modified this flpe by PCR:
1st PCR : cloning
-> ligation with vector
-> 2nd PCR : modified SpeI site
-> 3rd PCR : modified EcoRI site
We use this part as the form of reverse, so we made this part reverse by PCR.
Expression check
To check whether flpe works correctly, we made assay shown in Fig.
The top construct is a nagative control. GFP can't be expressed because of the double terminators.
The bottom construct express GFP when flpe works correctly. When flpe is expressed correctly, flpe recognaize the frt site and double terminator which terminates the expression of gfp is removed, so GFP may be expressed.
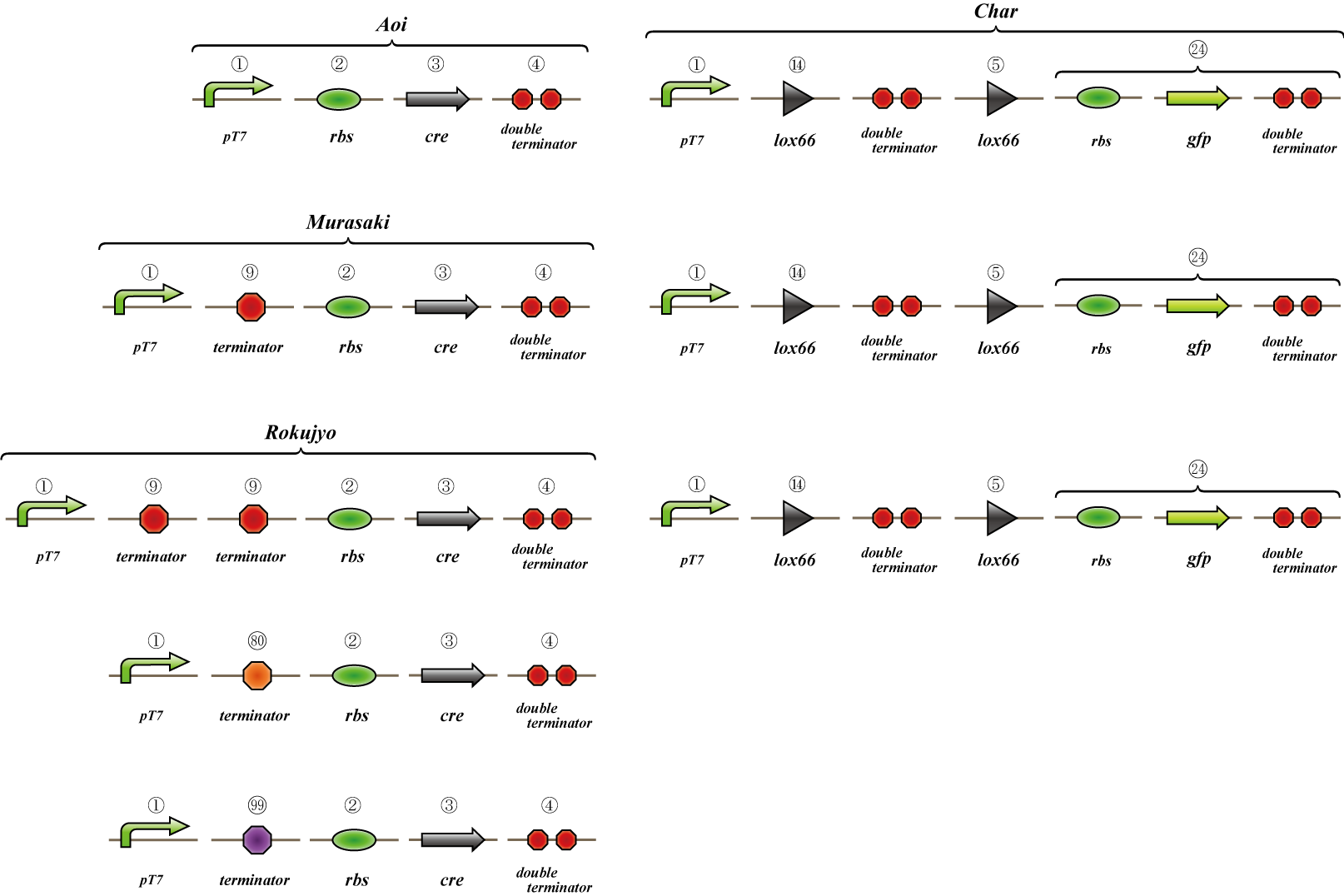
Parts list
| Number | Name | Link to BioBrick | Plate coordinate | Vector | Code length |
| 1 | T7 promoter |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_I712074 BBa_I712074] | plate1-6N | pSB1AK8 | 46bp |
| 2 | rbs |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_B0030 BBa_B0030] | plate1-1H | pSB1A2 | 15bp |
| 3 | cre recombinase |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J61047 BBa_J61047] | plate1-5D | pSB1A2 | 1037bp |
| 4 | double terminator |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 BBa_B0014] | plate2-24C | pSB1AK3 | 95bp |
| 5 | lox66 recombinase site |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_I718017 BBa_I718017] | plate1-17J | pSB1A2 | 34bp |
| 6 | Kan resistance (rev) |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J31002 BBa_J31002] | plate1-2K | pSB1A2 | 816bp |
| 7 | rbs (rev) |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J44001 BBa_J44001] | plate1-1J | pSB1A2 | 15bp |
| 8 | lox71 recombinase site |
order-made | --- | --- | 34bp |
| 9 | single terminator |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_B1006 BBa_B1006] | plate1-4H | pSB1AK3 | 39bp |
| 10 | constant express promoter |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J23119 BBa_J23119] | plate1-18A | pSB1A2 | 35bp |
| 11 | Tet resistance (rev) |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J31006 BBa_J31006] | plate1-1N | pSB1A2 | 1191bp |
| 12 | hixC |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J44000 BBa_J44000] | plate1-1B | pSB1A2 | 26bp |
| 13 | --- |
--- | --- | --- | --- |
| 14 | lox66 |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_I718016 BBa_I718016] | plate1-17H | pSB1A2 | 34bp |
| 15 | rbs-mRFP1-terminator |
[http://partsregistry.org/wiki/index.php?title=Part:BBa_I13507 BBa_I13507] | plate1-22O | pSB1A2 | 861bp |
| 16 | location sequence(old) |
order | --- | --- | --- |
| 17 | pSP6(rev) |
order | --- | --- | --- |
| 18 | lox2272 |
order | --- | --- | --- |
| 19 | loading sequence(rev) |
order | --- | --- | --- |
| 20 | frt |
[http://partsregistry.org/Part:BBa_J61020 BBa_J61020] | From HQ | pSB1A2 | 34bp |
| 21 | hin(true) |
[http://partsregistry.org/Part:BBa_J31000 BBa_J31000] | From HQ | ??? | 573bp |
| 22 | hin(rev) |
--- | --- | ??? | 573bp |
| 23 | flpe |
--- | --- | --- | about 1.2kbp |
| 24 | gfp unit |
--- | --- | --- | about 1.2kbp |
| 25 | flpe(rev) |
--- | --- | --- | about 1.2kbp |
[http://igem-ut.net/2010/now/ Latest Experiments]
Our Latest Experiment & results. Available from [http://igem-ut.net/2010/now/ here].
Detail Note Book
Detail protocols about Sudoku project are:
June.
July.
 "
"