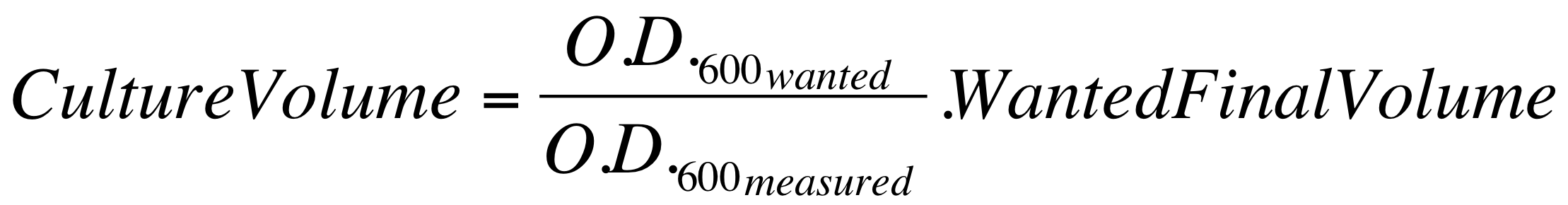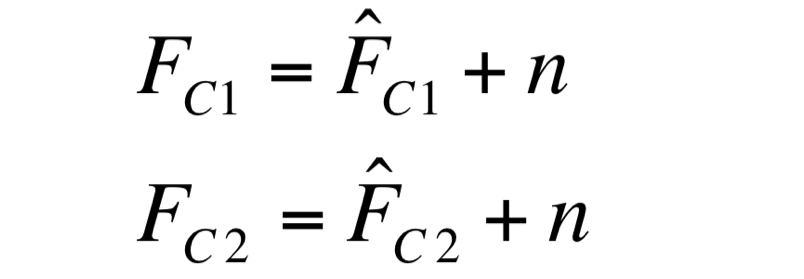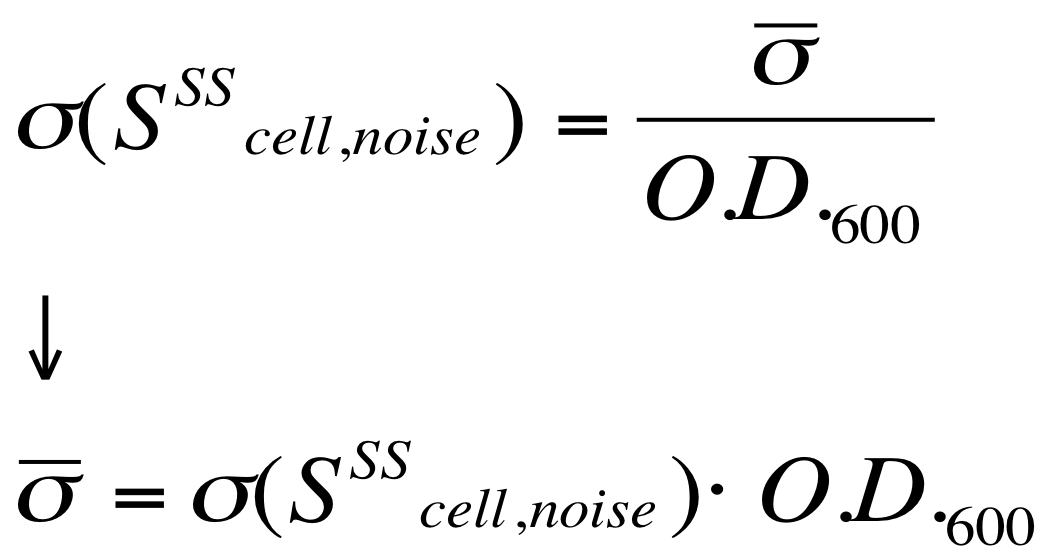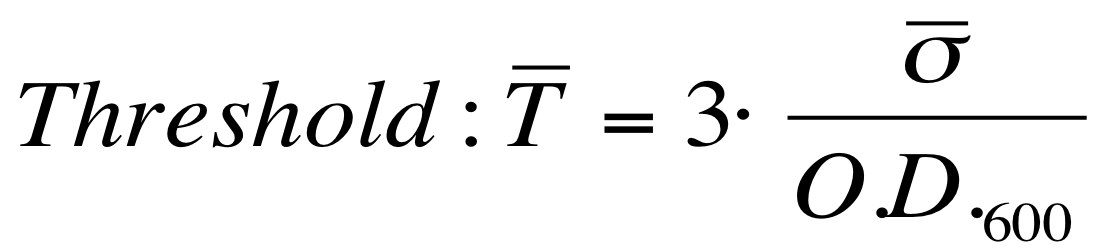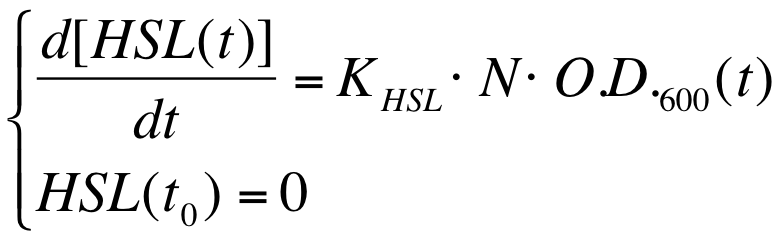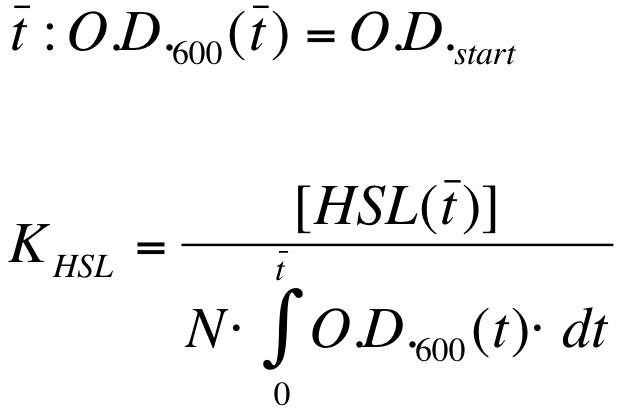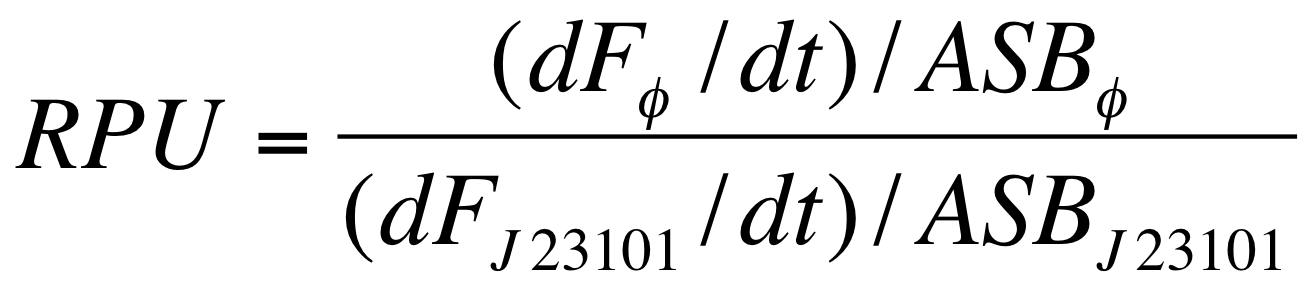Team:UNIPV-Pavia/Parts/Characterization
From 2010.igem.org
(→Data Analysis to estimate the HSL synthesys rate per cell) |
(→New Parts) |
||
| (22 intermediate revisions not shown) | |||
| Line 40: | Line 40: | ||
* [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300000 - BioBrick integrative base vector for E. coli |BBa_K300000 - BioBrick integrative base vector for E. coli]] | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300000 - BioBrick integrative base vector for E. coli |BBa_K300000 - BioBrick integrative base vector for E. coli]] | ||
* [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300001 - BioBrick integrative base vector for S. cerevisiae|BBa_K300001 - BioBrick integrative base vector for S. cerevisiae]] | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300001 - BioBrick integrative base vector for S. cerevisiae|BBa_K300001 - BioBrick integrative base vector for S. cerevisiae]] | ||
| - | |||
* [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300010 - PoPS-based self-inducible device|BBa_K300010 - PoPS-based self-inducible device]] | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300010 - PoPS-based self-inducible device|BBa_K300010 - PoPS-based self-inducible device]] | ||
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300093, BBa_K300094, BBa_K300097, BBa_K300095 and BBa_K300084 - Phasin and Intein-based tags for protein purification|BBa_K300093, BBa_K300094, BBa_K300097, BBa_K300095 and BBa_K300084 - Phasin and Intein-based tags for protein purification]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300093, BBa_K300094, BBa_K300097, BBa_K300095, BBa_K300086 and BBa_K300084 - Phasin and Intein-based tags for protein purification|BBa_K300093, BBa_K300094, BBa_K300097, BBa_K300095, BBa_K300086 and BBa_K300084 - Phasin and Intein-based tags for protein purification]] |
==[[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts |Improved Parts]]== | ==[[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts |Improved Parts]]== | ||
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300002 - Phasin (PhaP) - head domain|BBa_K300002 - Phasin (PhaP) - head domain]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300002 - Phasin (PhaP) - head domain|BBa_K300002 - Phasin (PhaP) - head domain - improvement of BBa_K208001]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300003 - Phasin (PhaP) - internal domain|BBa_K300003 - Phasin (PhaP) - internal domain]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300003 - Phasin (PhaP) - internal domain|BBa_K300003 - Phasin (PhaP) - internal domain - improvement of BBa_K208001]] |
| - | + | ||
==[[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry |Existing Parts from the Registry]]== | ==[[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry |Existing Parts from the Registry]]== | ||
| + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_R0010, BBa_R0011 - Wild type and hybrid lac promoters|BBa_R0010, BBa_R0011 - Wild type and hybrid lac promoters]] | ||
* [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device|BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device]] | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device|BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device]] | ||
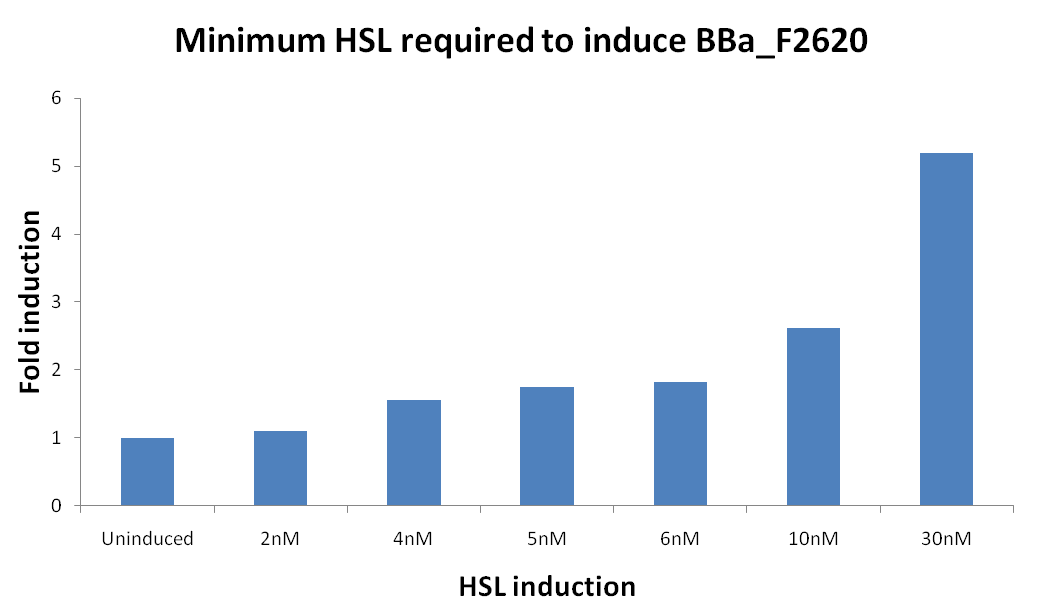
* [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_F2620 - 3OC6HSL -> PoPS Receiver|BBa_F2620 - 3OC6HSL -> PoPS Receiver]] | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_F2620 - 3OC6HSL -> PoPS Receiver|BBa_F2620 - 3OC6HSL -> PoPS Receiver]] | ||
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J61001 - Origin of replication|BBa_J61001 - Origin of replication]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J61001 - R6K Origin of replication|BBa_J61001 - R6K Origin of replication]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J23100, BBa_J23101, BBa_J23105, BBa_J23106, BBa_J23110, BBa_J23114, BBa_J23116, BBa_J23118 - constitutive Anderson | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J23100, BBa_J23101, BBa_J23105, BBa_J23106, BBa_J23110, BBa_J23114, BBa_J23116, BBa_J23118 - constitutive promoters from Anderson's collection|BBa_J23100, BBa_J23101, BBa_J23105, BBa_J23106, BBa_J23110, BBa_J23114, BBa_J23116, BBa_J23118 - constitutive promoters from Anderson's collection]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_P1004 - | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_P1004 - chloramphenicol resistance cassette|BBa_P1004 - chloramphenicol resistance cassette]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_K125500 - | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_K125500 - GFP fusion brick|BBa_K125500 - GFP fusion brick]] |
* [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J72008 - phi80 integration helper plasmid pInt80-649|BBa_J72008 - phi80 integration helper plasmid pInt80-649]] | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J72008 - phi80 integration helper plasmid pInt80-649|BBa_J72008 - phi80 integration helper plasmid pInt80-649]] | ||
<hr><br> | <hr><br> | ||
| Line 75: | Line 74: | ||
* 8 ul of long term storage glycerol stock were inoculated in 1 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | * 8 ul of long term storage glycerol stock were inoculated in 1 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
* The grown cultures were then diluted 1:100 in 1 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | * The grown cultures were then diluted 1:100 in 1 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | ||
| - | * | + | * Cultures were then pelletted (2000 rpm, 10 minutes) in order to eliminate the HSL produced during the growth. |
* Supernatants were discarded and the pellets were resuspended in 1 ml of LB or M9 + suitable antibiotic and transferred to a 1.5 ml tube | * Supernatants were discarded and the pellets were resuspended in 1 ml of LB or M9 + suitable antibiotic and transferred to a 1.5 ml tube | ||
| - | * These cultures were diluted 1:1000 in 1 ml of LB or M9 + suitable antibiotic and aliquoted in a flat-bottom 96-well microplate in triplicate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects | + | * These cultures were diluted 1:1000 in 1 ml of LB or M9 + suitable antibiotic and aliquoted in a flat-bottom 96-well microplate in triplicate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects, see https://2009.igem.org/Team:UNIPV-Pavia/Methods_Materials/Evaporation). All the wells were filled with a 200 ul volume. |
* The microplate was incubated in the Tecan Infinite F200 microplate reader and fluorescence and absorbance were measured with this automatic protocol: | * The microplate was incubated in the Tecan Infinite F200 microplate reader and fluorescence and absorbance were measured with this automatic protocol: | ||
** 37°C constant for all the experiment; | ** 37°C constant for all the experiment; | ||
| Line 96: | Line 95: | ||
*8 ul of long term storage glycerol stock were inoculated in 5 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | *8 ul of long term storage glycerol stock were inoculated in 5 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
*The grown cultures were then diluted 1:100 in 5 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | *The grown cultures were then diluted 1:100 in 5 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | ||
| - | *These new cultures were diluted to an O.D.600 of 0.02 (measured with a TECAN F200 microplate reader on a 200 ul of volume per well; it is not the 1 cm pathlength cuvette) in 2 ml LB or M9 + suitable antibiotic. In order to have the cultures at the desired O.D.600, the following dilution was performed: | + | *These new cultures were diluted to an O.D.600 of 0.02 (measured with a TECAN F200 microplate reader on a 200 ul of volume per well; it is not equivalent to the 1 cm pathlength cuvette) in 2 ml (wanted final volume) LB or M9 + suitable antibiotic. In order to have the cultures at the desired O.D.600 (O.D._wanted=0.02), the following dilution was performed: |
[[Image:UNIPV_Pavia_OD600_dil.png|500px|center]] | [[Image:UNIPV_Pavia_OD600_dil.png|500px|center]] | ||
*These new dilutions were aliquoted in a flat-bottom 96-well microplate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects in the frame). All the wells were filled with a 200 ul volume. | *These new dilutions were aliquoted in a flat-bottom 96-well microplate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects in the frame). All the wells were filled with a 200 ul volume. | ||
| Line 122: | Line 121: | ||
**8 ul of BBa_J231xx-<partinfo>BBa_K300009</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | **8 ul of BBa_J231xx-<partinfo>BBa_K300009</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
**The grown cultures were then diluted 1:100 in 5 ml of LB supplemented medium and incubated in the same conditions as before for 6 hours. | **The grown cultures were then diluted 1:100 in 5 ml of LB supplemented medium and incubated in the same conditions as before for 6 hours. | ||
| - | ** | + | **Each falcon was pelletted (2000 rpm, 10 minutes) and supernatants were collected and filtered (0.2 um), in order to eliminate bacterial residues. These supernatants contained the 3OC6-HSL at the concentration produced by cultures. These supernatants were used to "induce" <partinfo>BBa_T9002</partinfo> cultures, in order to quantify the [HSL]. They were conserved at -20°C until the next day. |
| - | + | ||
*<em>Growth and preparation of the biosensor culture of <partinfo>BBa_T9002</partinfo></em> | *<em>Growth and preparation of the biosensor culture of <partinfo>BBa_T9002</partinfo></em> | ||
**8 ul of <partinfo>BBa_T9002</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | **8 ul of <partinfo>BBa_T9002</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
| Line 141: | Line 139: | ||
*** 0,1 nM | *** 0,1 nM | ||
*** 0 M | *** 0 M | ||
| - | **All inductions were performed adding 2ul of "inducer solution" (3OC6-HSL, Sigma Aldrich) at | + | **All inductions were performed adding 2ul of "inducer solution" (3OC6-HSL, Sigma Aldrich) at the proper concentration in 200 ul of culture. |
*<em>HSL quantification</em> | *<em>HSL quantification</em> | ||
**2ul of supernatants prepared as described above were used to induce 200 ul of <partinfo>BBa_T9002</partinfo> cultures in triplicate. | **2ul of supernatants prepared as described above were used to induce 200 ul of <partinfo>BBa_T9002</partinfo> cultures in triplicate. | ||
| - | **If the amount of 3OC6-HSL present in the supernatant was not enough concentrated to trigger the induction of <partinfo>BBa_T9002</partinfo>, a bigger amount of supernatant (X ul) was used to induce the cultures (200-X ul of cultures). | + | **If the amount of 3OC6-HSL present in the supernatant was not enough concentrated to trigger the induction of <partinfo>BBa_T9002</partinfo>, a bigger amount of supernatant (X ul) was used to induce the cultures (200-X ul of cultures). Then, the calibration curve was obtained by using the same amount of <partinfo>BBa_T9002</partinfo> culture (200-X ul) induced with X ul of inducer solution at the proper concentration. To maintain the same experimental conditions, the inducer was diluted in the supernatant of a negative control (i.e., <partinfo>BBa_B0034</partinfo>). |
**Fluorescence and absorbance were measured after 30 min from the induction with a Tecan Infinite F200 microplate reader and, using the information provided by the calibration curve, the amount of 3OC6-HSL present in every supernatant was evaluated (each reported fluorescence value was corrected with the O.D.600 of the culture). | **Fluorescence and absorbance were measured after 30 min from the induction with a Tecan Infinite F200 microplate reader and, using the information provided by the calibration curve, the amount of 3OC6-HSL present in every supernatant was evaluated (each reported fluorescence value was corrected with the O.D.600 of the culture). | ||
| Line 151: | Line 149: | ||
---- | ---- | ||
<br> | <br> | ||
| - | |||
===Microplate reader experiments for <partinfo>BBa_F2620</partinfo> - Protocol #4=== | ===Microplate reader experiments for <partinfo>BBa_F2620</partinfo> - Protocol #4=== | ||
| Line 166: | Line 163: | ||
<br> | <br> | ||
===Preliminary remarks=== | ===Preliminary remarks=== | ||
| - | *All our growth curves have been obtained subtracting for each time sample the broth O.D.600 measurement from that of the culture; broth was considered in the same conditions of the culture ( | + | *All our growth curves have been obtained subtracting for each time sample the broth O.D.600 measurement from that of the culture; broth was considered in the same conditions of the culture (i.e. induced with the same inducer concentration and supplemented with the same antibiotic of the culture). |
| - | *Fluorescence signals have been obtained subtracting for each time sample the fluorescent measurement of a non-fluorescent culture from that of the target culture. The non-fluorescent culture was considered in the same conditions of the | + | *Fluorescence signals have been obtained subtracting for each time sample the fluorescent measurement of a non-fluorescent culture from that of the target culture. The non-fluorescent culture was considered in the same conditions of the culture of interest (e.g. induced with the same inducer concentration and with the same plasmid/antibiotic resistance, but without fluorescent reporter genes). This operation allows the removal, from the target fluorescent signal, of the "self-fluorescent" component and the fluorescence signal obtained is "blanked". |
| Line 193: | Line 190: | ||
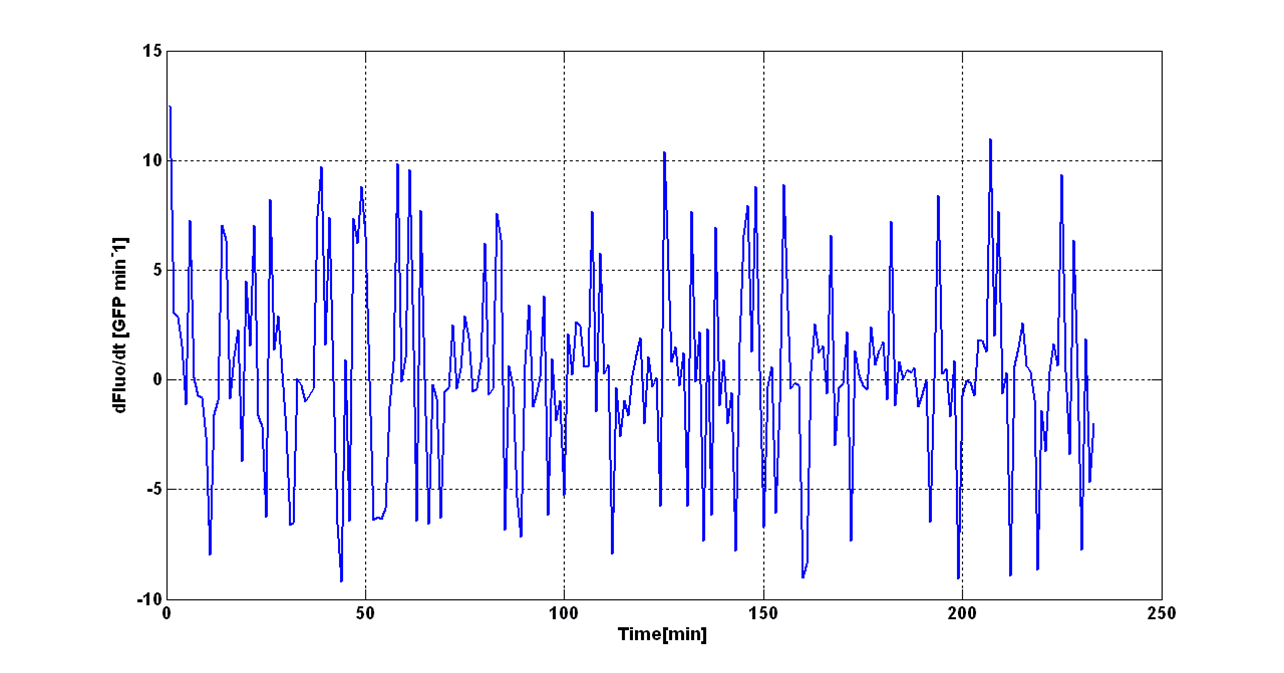
O.D.start was evaluated by computing the ''Scell'' (GFPmut3 synthesis rate per cell) signal for the desired self-inducible promoter. | O.D.start was evaluated by computing the ''Scell'' (GFPmut3 synthesis rate per cell) signal for the desired self-inducible promoter. | ||
| - | Scell was obtained by computing (1/O.D.600)*dGFP/dt, where O.D.600 and GFP are the | + | Scell was obtained by computing (1/O.D.600)*dGFP/dt, where O.D.600 and GFP are the blanked absorbance and fluorescence signals. |
The goal is the estimation of the critical O.D.600 value (O.D.start) at which the Scell significantly increases. Because Scell is a very noisy signal, a threshold value which takes into account the noise variability was proposed and it is described below. | The goal is the estimation of the critical O.D.600 value (O.D.start) at which the Scell significantly increases. Because Scell is a very noisy signal, a threshold value which takes into account the noise variability was proposed and it is described below. | ||
| Line 199: | Line 196: | ||
Two different signals, measured from independent samples of the same non-fluorescent culture in the same experiment, are considered: C1 and C2. The fluorescence signal of C1 and C2 (F_C1 and F_C2 respectively) can be thought as the addition of a “real signal” and a noise component. | Two different signals, measured from independent samples of the same non-fluorescent culture in the same experiment, are considered: C1 and C2. The fluorescence signal of C1 and C2 (F_C1 and F_C2 respectively) can be thought as the addition of a “real signal” and a noise component. | ||
| - | [[Image:UNIPV_Pavia_noise1.png| | + | [[Image:UNIPV_Pavia_noise1.png|200px|center]] |
F_C1 and F_C2 have the same expected value and the same standard deviation, since they are two independent realizations of the same aleatory process: in fact, they are two time series acquired from the same cultures in the same growth conditions by the same instrument. | F_C1 and F_C2 have the same expected value and the same standard deviation, since they are two independent realizations of the same aleatory process: in fact, they are two time series acquired from the same cultures in the same growth conditions by the same instrument. | ||
| Line 244: | Line 241: | ||
<br> | <br> | ||
| - | ===Data | + | ===Data analysis to estimate the HSL synthesis rate per cell=== |
The autoinducer synthesys rate per cell is a very important parameter to be evaluated in quorum sensing systems. | The autoinducer synthesys rate per cell is a very important parameter to be evaluated in quorum sensing systems. | ||
| - | In fact the knowledge of this parameter enables the rational design of cell-communication systems, such as the self inducible devices studied in this project. | + | In fact the knowledge of this parameter enables the rational design of cell-communication systems, such as the self-inducible devices studied in this project. |
A model based approach was proposed to estimate this interesting parameter. | A model based approach was proposed to estimate this interesting parameter. | ||
| Line 272: | Line 269: | ||
|[[Image:PV_Immagine_formula2.png|300px]] | |[[Image:PV_Immagine_formula2.png|300px]] | ||
|} | |} | ||
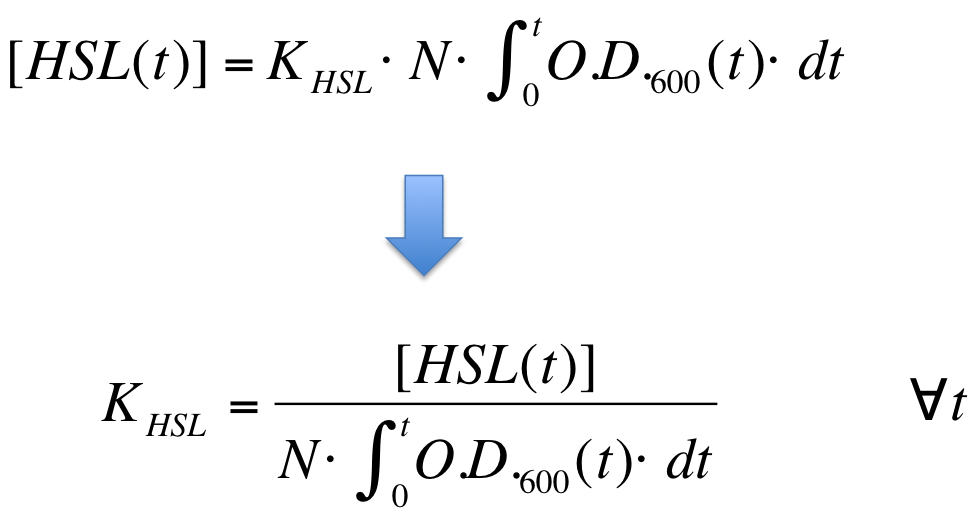
| - | K_HSL can be estimated for t=t_bar, corresponding to the | + | K_HSL can be estimated for t=t_bar, corresponding to the transcription initiation time, as reported below. |
{| align='center' | {| align='center' | ||
| Line 279: | Line 276: | ||
where: | where: | ||
* N was estimated by counting TOP10 colonies obtained by plating serial dilutions of cultures with known O.D.600. | * N was estimated by counting TOP10 colonies obtained by plating serial dilutions of cultures with known O.D.600. | ||
| - | * [HSL(t_bar)] was estimated as described in the section [[Data Analysis - minimum induction required to activate ''lux pR'' for <partinfo>BBa_F2620</partinfo>]] | + | * [HSL(t_bar)] was estimated as described in the section [[Team:UNIPV-Pavia/Parts/Characterization#Data Analysis - minimum induction required to activate lux pR for BBa_F2620|Data analysis - minimum induction required to activate ''lux pR'' for <partinfo>BBa_F2620</partinfo>]] |
* the O.D.600 time series was measured in each experiment, so its time integral can be computed by trapezoidal numerical integration. | * the O.D.600 time series was measured in each experiment, so its time integral can be computed by trapezoidal numerical integration. | ||
| - | N was estimated and values are reported below: | + | N was estimated in LB and M9 media and values are reported below: |
{| border='1' align='center' | {| border='1' align='center' | ||
| Line 321: | Line 318: | ||
===Data analysis for RPU evaluation=== | ===Data analysis for RPU evaluation=== | ||
| - | The RPUs are standard units proposed by Kelly J. et al., | + | The RPUs are standard units proposed by Kelly J. et al., 2009, in which the relative transcriptional strength of a promoter can be measured using a reference standard. |
RPUs have been computed as: | RPUs have been computed as: | ||
Latest revision as of 02:42, 28 October 2010
|
|
|||||||||||||||||||||||||
|
|
||||||||||||||||||||||||
 "
"