Team:NYMU-Taipei/Experiments/Speedy switch
From 2010.igem.org
(→Repoting Assay 4) |
|||
| (109 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:NYMU-Taipei/Header}} | ||
| + | |||
=Method = | =Method = | ||
| - | *Protocol: | + | *Protocol: (Harbaugh S.V., 2008) |
1.Selected genes to be reported are incubated overnight in an LB liquid culture at 37oC and 180-200rpm. This makes sure they are fresh in the morning. Positive and negative controls are also needed. | 1.Selected genes to be reported are incubated overnight in an LB liquid culture at 37oC and 180-200rpm. This makes sure they are fresh in the morning. Positive and negative controls are also needed. | ||
| Line 14: | Line 16: | ||
5.Measurement of OD at 4 hours: For each used well in the 96-well plate: | 5.Measurement of OD at 4 hours: For each used well in the 96-well plate: | ||
| - | Take the liquid from the well and put it in the cuvette to measure the OD ["OD at 4 hours"]. | + | Take the liquid from the well and put it in the cuvette to measure the OD ["OD at 4 hours"]. |
| + | |||
| + | *The optimizing data: | ||
| + | We took the data from OD600 of each sample, which should have an exponential growth curve, and took the ln of each value. After taking the logarithm of the data, we created a linear curve. Since we have the two end points of the OD 600 of each sample, we use this linear curve to modulate OD value of each sample at each specific time point. This value was then recalculated back into its original curve using exponents. | ||
| + | Our fluorescent data was normalized by taking the fluorescence of our sample at each time point and subtracting the fluorescence of the negative control in the same OD value at the same time point. | ||
| + | Finally, plot the normalized fluorescence versus time in minutes scale. | ||
| + | |||
| + | =Reporting Assay= | ||
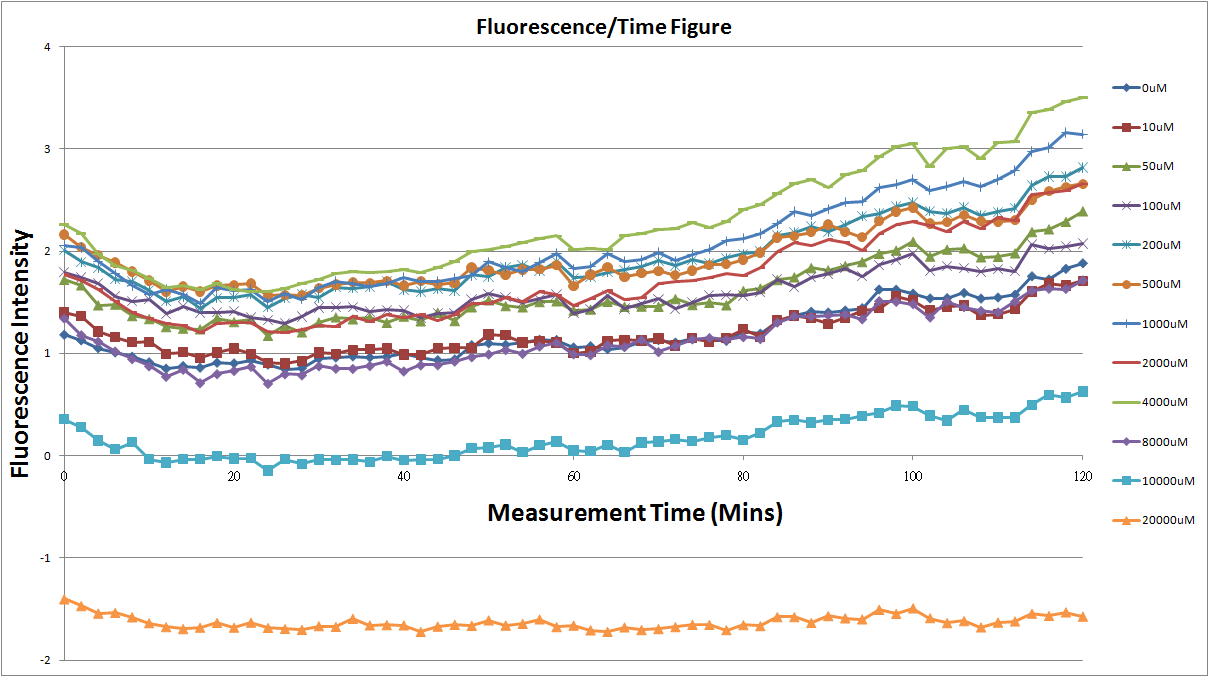
| + | *Fig.1, fig.3, fig.5, and fig.7 are the original charts of the experiment. N line and NT line are control line. N line stands for the cell which sequence doesn't have pLac promoter.NT line stands for the cell which sequence doesn't have promoter but added 0.1mM Theophylline. N line shows that even though the sequence doesn't have promoter it still have little fluorescence so we use it to modify the instrument errors. 0μM sample which sequence has promoter but doesn’t added Theophylline still has little fluorescence because mRNA is leak. Normalizing with the N, we use this line to find out what's the degree of mRNA leak. NT line which doesn’t have promoter in the plasmid but add 0.1 mM Theophylline still is detected fluorescence. We added Theophylline in DMSO, so we use this NT line to normalize the other samples which added different concentration of Theophylline in order to eliminate the spectrum effect of Theophylline. | ||
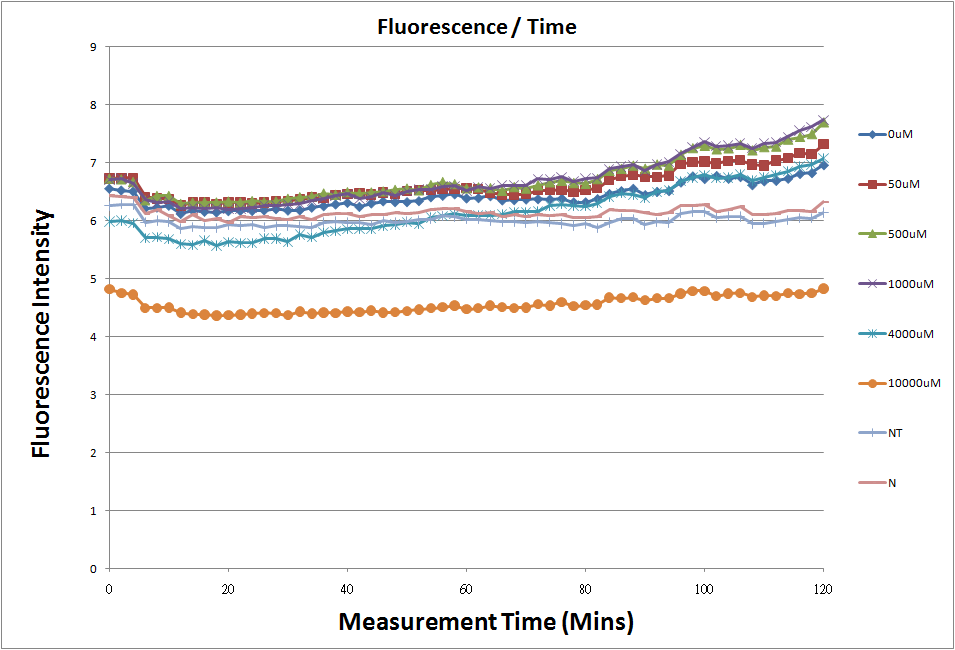
| + | *Fig.2, fig.4, fig.6, and fig.8 are charts which have been normalized with the control N line and NT line. | ||
| + | |||
| + | ==Reporting Assay 1== | ||
| + | *[https://2010.igem.org/Team:NYMU-Taipei/Experiments/Riboswitch#2010.10.22 Reagent formula] | ||
| + | |||
| + | |||
| + | * The oringinal fluorescence data. | ||
| + | [[Image:20101023(re).png|thumb|none|800px|Fig.1: The 12 different concentration of theophylline and control group comparision.]] | ||
| + | |||
| + | * Normalized the 12 different Theophylline concentration samples with N. | ||
| + | [[Image:Plot20101023.png|thumb|none|800px|Fig.2: The 12 different concentration of theophylline comparision.]] | ||
| + | |||
| + | *Discussion | ||
| + | **Under 4mM Theophylline , we added more concentrations of theophylline the protein expressed stronger ,so we can see the higher fluorescent intensity. | ||
| + | **The 8mM and 10mM lines were low initially and then growing.We sugested that may have two possibility.One is that because we take out ''Escherichia coli'' from 37 degrees centigrade and added theophylline in it at room temperature and we take the ''Escherichia coli'' into 37 degrees centigrade incubator.So the ''Escherichia coli'' may not get with the temperature and die initially. | ||
| + | **The 20mM line show that the theophylline concentration reached the ''Escherichia coli'' limitation and ''Escherichia coli'' was dead. | ||
| + | |||
| + | ==Reporting Assay 2== | ||
| + | *[https://2010.igem.org/Team:NYMU-Taipei/Experiments/Riboswitch#2010.10.24 Reagent Formula] | ||
| + | |||
| + | *The original fluorescence data. | ||
| + | [[Image:20101024(org).png|thumb|none|800px|Fig.3:The 12 different concentration of theophylline and control group comparision. ]] | ||
| + | *Normalized 12 different concentration of Theophylline with NT. | ||
| + | [[Image:20101024.png|thumb|none|800px|Fig.4:The 12 different concentration of theophylline comparision. ]] | ||
| + | *Discussion | ||
| + | **Under 8mM theophylline,we added more concentrations of theophylline the protein expressed strongers so we can see the higher fluorescent intensity. | ||
| + | **The 10mM lines was low initially and then growing.We sugested that may have two possibility.One is that because we take out ''Escherichia coli'' from 37 degrees centigrade and added theophylline in it at room temperature and we take the ''Escherichia coli'' into 37 degrees centigrade incubator.So the ''Escherichia coli'' may not get with the temperature and die initially. | ||
| + | **The 20mM line show that the theophylline concentration reached the ''Escherichia coli'' limitation and ''Escherichia coli'' was dead. | ||
| + | |||
| + | ==Reporting Assay 3== | ||
| + | *[https://2010.igem.org/Team:NYMU-Taipei/Experiments/Riboswitch#2010.10.25 Reagent formula] | ||
| + | |||
| + | *The oringinal fluorescence data. | ||
| + | [[Image:20101025(org).png|thumb|none|800px|Fig.5:The 6 different concentration of theophylline and control group comparision. ]] | ||
| + | |||
| + | *Normalized the 6 different Theophylline concentration samples with NT & N. | ||
| + | [[Image:20101025.png|thumb|none|800px|Fig.6:The 6 different concentration of theophylline comparision. ]] | ||
| + | |||
| + | *Discussion | ||
| + | **Under 4mM theophylline,we added more concentrations of theophylline the protein expressed strongers so we can see the higher fluorescent intensity .In the assay 3 & 4, we revised the reagent formula in order to eliminate the total volume effect of the sample because the volume of Theophylline we add should be considered. As a result, we modify the LB liquid volume to fit the total volume of each sample into 4mL. By this modifying method, we can make sure the concentration of Theophylline in each sample. Comparing to the assay1 & 2, we can find that 4mM sample is still above the 0 mM curve because the real concentration of this sample is below 4mM. Also, we can find that 4 mM sample is invalid. | ||
| + | **The 10mM lines was low initially and then growing.We sugested that may have two possibility.One is that because we take out ''Escherichia coli'' from 37 degrees centigrade and added theophylline in it at room temperature and we take the ''Escherichia coli'' into 37 degrees centigrade incubator.So the ''Escherichia coli'' may not get with the temperature and die initially. | ||
| + | |||
| + | ==Repoting Assay 4== | ||
| + | *[https://2010.igem.org/Team:NYMU-Taipei/Experiments/Riboswitch#2010.10.26 Reagent Formula] | ||
| + | *The original fluorescence data. | ||
| + | [[Image:20101026(org).png|thumb|none|800px|Fig.7:The 6 different concentration of theophylline and control group comparision. ]] | ||
| + | |||
| + | *Normalized 6 different concentration of Theophylline with NT & N. | ||
| + | [[Image:20101026.png|thumb|none|800px|Fig.7:The 6 different concentration of theophylline comparision. ]] | ||
| + | *Discussion | ||
| + | **Under 4mM theophylline,we added more concentrations of theophylline the protein expressed strongers so we can see the higher fluorescent intensity . | ||
| + | **The 10mM line show that the theophylline concentration reached the ''Escherichia coli'' limitation and ''Escherichia coli'' was dead. | ||
| - | + | =Conclusion= | |
| - | = | + | * When the concentration of Theophylline is in the range 0.01mM to 2mM, the concentration of Theophylline and the resulting fluorescence (using the {{:Team:NYMU-Taipei/BBa|K411003}} construct) are directly proportional. As a result, we recommend the Theophylline riboswitch ({{:Team:NYMU-Taipei/BBa|K411001}}) to be used in that range to control the downstream translation. |
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | * | + | * The study by Suess ''et al'', adding more than 5mM of Theophylline would cause ''E. coli'' to die (Suess, 2004). In our experiments, we find that after adding more than 4mM, the Theophylline spectrum curve would be invalid. As a result, we do not recommend doing experiments with concentrations over 4mM as the ''E. coli'' cell would be unstable or the regulation of the riboswitch would not be accurate. |
| - | + | ||
| - | + | * Our figures show that the sample fluorescence will disperse clearly after 80 minutes. As a result, we suggest that the protein expression of the downstream coding sequence will be more obvious after about 200 minutes of adding Theophylline. | |
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | =Reference= |
| - | + | *Beatrix Suess, Barbara Fink, Christian Berens, ReÂgis Stentz and Wolfgang Hillen(2004)A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Research, Vol. 32, No. 4 ) | |
| + | *Harbaugh S.V., Davidson M.E., Chushak Y.G., Kelley-Loughnane N., and Stone M.O.(2008) Riboswitch-based sensor in low optical background. Proc. of SPIE 7040, 70400C | ||
| - | |||
| - | |||
| - | + | {{:Team:NYMU-Taipei/Footer}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 02:22, 28 October 2010
| Home | Project Overview | Speedy reporter | Speedy switch | Speedy protein degrader | Experiments and Parts | Applications | F.A.Q | About Us |
Contents |
Method
- Protocol: (Harbaugh S.V., 2008)
1.Selected genes to be reported are incubated overnight in an LB liquid culture at 37oC and 180-200rpm. This makes sure they are fresh in the morning. Positive and negative controls are also needed.
2.Overnight liquid culture is diluted to OD600 of 0.1, Theophylline is added at concentrations ranging from 0.01mM to 20mM, and the mix incubated for 2-2.5 hours.
3.Measurement of OD at 2 hours: For each used well in the 96-well plate: Take 200uL from the liquid (make sure you pipette this step well) and put it in a cuvette to read the OD600. Note down the OD600 ["OD at 2 hours"], then take the liquid in the cuvette and put it in the right place in the 96-well plate.
4.Measurement of fluorescence: Continuous measurement of fluorescence with the excitation/emission wavelengths 488/511nm for 2 hours, with one fluorescence data point every 2 minutes.
5.Measurement of OD at 4 hours: For each used well in the 96-well plate: Take the liquid from the well and put it in the cuvette to measure the OD ["OD at 4 hours"].
- The optimizing data:
We took the data from OD600 of each sample, which should have an exponential growth curve, and took the ln of each value. After taking the logarithm of the data, we created a linear curve. Since we have the two end points of the OD 600 of each sample, we use this linear curve to modulate OD value of each sample at each specific time point. This value was then recalculated back into its original curve using exponents. Our fluorescent data was normalized by taking the fluorescence of our sample at each time point and subtracting the fluorescence of the negative control in the same OD value at the same time point. Finally, plot the normalized fluorescence versus time in minutes scale.
Reporting Assay
- Fig.1, fig.3, fig.5, and fig.7 are the original charts of the experiment. N line and NT line are control line. N line stands for the cell which sequence doesn't have pLac promoter.NT line stands for the cell which sequence doesn't have promoter but added 0.1mM Theophylline. N line shows that even though the sequence doesn't have promoter it still have little fluorescence so we use it to modify the instrument errors. 0μM sample which sequence has promoter but doesn’t added Theophylline still has little fluorescence because mRNA is leak. Normalizing with the N, we use this line to find out what's the degree of mRNA leak. NT line which doesn’t have promoter in the plasmid but add 0.1 mM Theophylline still is detected fluorescence. We added Theophylline in DMSO, so we use this NT line to normalize the other samples which added different concentration of Theophylline in order to eliminate the spectrum effect of Theophylline.
- Fig.2, fig.4, fig.6, and fig.8 are charts which have been normalized with the control N line and NT line.
Reporting Assay 1
- The oringinal fluorescence data.
- Normalized the 12 different Theophylline concentration samples with N.
- Discussion
- Under 4mM Theophylline , we added more concentrations of theophylline the protein expressed stronger ,so we can see the higher fluorescent intensity.
- The 8mM and 10mM lines were low initially and then growing.We sugested that may have two possibility.One is that because we take out Escherichia coli from 37 degrees centigrade and added theophylline in it at room temperature and we take the Escherichia coli into 37 degrees centigrade incubator.So the Escherichia coli may not get with the temperature and die initially.
- The 20mM line show that the theophylline concentration reached the Escherichia coli limitation and Escherichia coli was dead.
Reporting Assay 2
- The original fluorescence data.
- Normalized 12 different concentration of Theophylline with NT.
- Discussion
- Under 8mM theophylline,we added more concentrations of theophylline the protein expressed strongers so we can see the higher fluorescent intensity.
- The 10mM lines was low initially and then growing.We sugested that may have two possibility.One is that because we take out Escherichia coli from 37 degrees centigrade and added theophylline in it at room temperature and we take the Escherichia coli into 37 degrees centigrade incubator.So the Escherichia coli may not get with the temperature and die initially.
- The 20mM line show that the theophylline concentration reached the Escherichia coli limitation and Escherichia coli was dead.
Reporting Assay 3
- The oringinal fluorescence data.
- Normalized the 6 different Theophylline concentration samples with NT & N.
- Discussion
- Under 4mM theophylline,we added more concentrations of theophylline the protein expressed strongers so we can see the higher fluorescent intensity .In the assay 3 & 4, we revised the reagent formula in order to eliminate the total volume effect of the sample because the volume of Theophylline we add should be considered. As a result, we modify the LB liquid volume to fit the total volume of each sample into 4mL. By this modifying method, we can make sure the concentration of Theophylline in each sample. Comparing to the assay1 & 2, we can find that 4mM sample is still above the 0 mM curve because the real concentration of this sample is below 4mM. Also, we can find that 4 mM sample is invalid.
- The 10mM lines was low initially and then growing.We sugested that may have two possibility.One is that because we take out Escherichia coli from 37 degrees centigrade and added theophylline in it at room temperature and we take the Escherichia coli into 37 degrees centigrade incubator.So the Escherichia coli may not get with the temperature and die initially.
Repoting Assay 4
- Reagent Formula
- The original fluorescence data.
- Normalized 6 different concentration of Theophylline with NT & N.
- Discussion
- Under 4mM theophylline,we added more concentrations of theophylline the protein expressed strongers so we can see the higher fluorescent intensity .
- The 10mM line show that the theophylline concentration reached the Escherichia coli limitation and Escherichia coli was dead.
Conclusion
- When the concentration of Theophylline is in the range 0.01mM to 2mM, the concentration of Theophylline and the resulting fluorescence (using the [http://partsregistry.org/Part:BBa_K411003 BBa_K411003] construct) are directly proportional. As a result, we recommend the Theophylline riboswitch ([http://partsregistry.org/Part:BBa_K411001 BBa_K411001]) to be used in that range to control the downstream translation.
- The study by Suess et al, adding more than 5mM of Theophylline would cause E. coli to die (Suess, 2004). In our experiments, we find that after adding more than 4mM, the Theophylline spectrum curve would be invalid. As a result, we do not recommend doing experiments with concentrations over 4mM as the E. coli cell would be unstable or the regulation of the riboswitch would not be accurate.
- Our figures show that the sample fluorescence will disperse clearly after 80 minutes. As a result, we suggest that the protein expression of the downstream coding sequence will be more obvious after about 200 minutes of adding Theophylline.
Reference
- Beatrix Suess, Barbara Fink, Christian Berens, ReÂgis Stentz and Wolfgang Hillen(2004)A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Research, Vol. 32, No. 4 )
- Harbaugh S.V., Davidson M.E., Chushak Y.G., Kelley-Loughnane N., and Stone M.O.(2008) Riboswitch-based sensor in low optical background. Proc. of SPIE 7040, 70400C
 "
"