Team:UNIPV-Pavia/Project/PromotoriAuto
From 2010.igem.org
(→Modulation of plasmid copy number) |
(→Results) |
||
| (14 intermediate revisions not shown) | |||
| Line 110: | Line 110: | ||
*high copy number plasmids and M9; | *high copy number plasmids and M9; | ||
*low copy number plasmids and M9. | *low copy number plasmids and M9. | ||
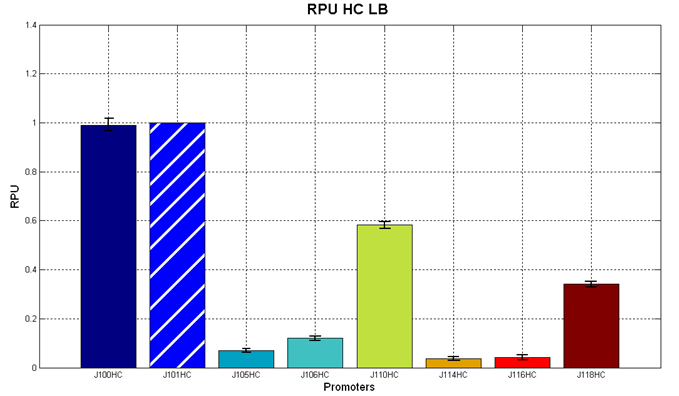
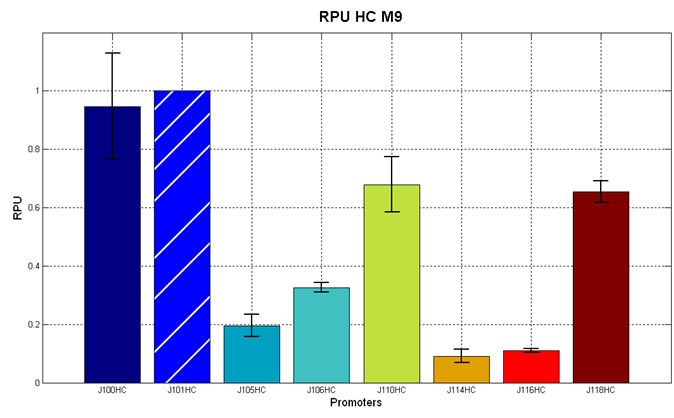
| - | It was not possible to evaluate promoters activities in low copy number plasmids in LB because the RFP activity was too weak and not distinguishable from the background. RFP fluorescence and Optical Density at 600nm (O.D.600) were measured in 96-well microplates, as reported in [[Team:UNIPV-Pavia/Parts/Characterization#Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2|Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2]] | + | It was not possible to evaluate promoters activities in low copy number plasmids in LB because the RFP activity was too weak and not distinguishable from the background. RFP fluorescence and Optical Density at 600nm (O.D.600) were measured in 96-well microplates, as reported in [[Team:UNIPV-Pavia/Parts/Characterization#Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2|Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2]]. Data were analyzed as reported in [[Team:UNIPV-Pavia/Parts/Characterization#Data analysis for RPU evaluation|Data analysis for RPU evaluation]]; |
'''Results''': results are shown here. | '''Results''': results are shown here. | ||
| Line 118: | Line 118: | ||
|} | |} | ||
{| align='center' | {| align='center' | ||
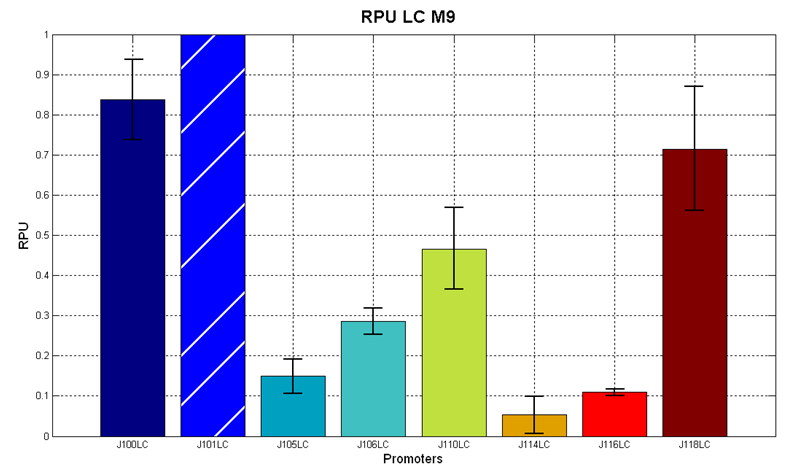
| - | |[[Image:pv_RPU_LC_M9.png|330px|thumb|center|Figure 7 - R.P.U. of the studied promoters from Anderson promoters' collection, M9 medium and low copy plasmid (<partinfo>pSB4C5</partinfo>). These plasmids were constructed by assembling the EcoRI-PstI | + | |[[Image:pv_RPU_LC_M9.png|330px|thumb|center|Figure 7 - R.P.U. of the studied promoters from Anderson promoters' collection, M9 medium and low copy plasmid (<partinfo>pSB4C5</partinfo>). These plasmids were constructed by assembling the EcoRI-PstI fragment of <partinfo>BBa_J61002</partinfo>-BBa_J231xx in <partinfo>pSB4C5</partinfo> vector, in order to transfer the promoter and the RBS-RFP-TT expression construct from <partinfo>BBa_J61002</partinfo> to <partinfo>pSB4C5</partinfo>.]] |
|} | |} | ||
| - | Doubling times were evaluated for the described cultures: | + | Doubling times were evaluated for the described cultures (HC stands for High Copy and LC stands for Low Copy): |
{| align='center' border='1' | {| align='center' border='1' | ||
|rowspan='2'|<b>Promoter</b> | |rowspan='2'|<b>Promoter</b> | ||
| Line 128: | Line 128: | ||
| LB in HC plasmid || M9 in HC plasmid || M9 in LC plasmid | | LB in HC plasmid || M9 in HC plasmid || M9 in LC plasmid | ||
|- | |- | ||
| - | |<partinfo>BBa_J23100</partinfo> || || || | + | |<partinfo>BBa_J23100</partinfo> || 33.75 <br> ± <br> 1.34 || 82.53 <br> ± <br> 2.45 || 86.11 <br> ± <br> 4.45 |
|- | |- | ||
| - | |<partinfo>BBa_J23101</partinfo>|| || || | + | |<partinfo>BBa_J23101</partinfo> || 35.93 <br> ± <br> 0.62 || 82.68 <br> ± <br> 1.84 || 86.42 <br> ± <br> 1.91 |
|- | |- | ||
| - | |<partinfo>BBa_J23105</partinfo>|| || || | + | |<partinfo>BBa_J23105</partinfo> || 29.86 <br> ± <br> 0.33 || 63.09 <br> ± <br> 7.08 || 85.00 <br> ± <br> 5.13 |
|- | |- | ||
| - | |<partinfo>BBa_J23106</partinfo>|| || || | + | |<partinfo>BBa_J23106</partinfo> || 29.17 <br> ± <br> 0.96 || 68.11 <br> ± <br> 4.25 || 88.71 <br> ± <br> 0.90 |
|- | |- | ||
| - | |<partinfo>BBa_J23110</partinfo>|| || || | + | |<partinfo>BBa_J23110</partinfo> || 31.28 <br> ± <br> 0.42 || 67.52 <br> ± <br> 5.87 || 76.15 <br> ± <br> 2.16 |
|- | |- | ||
| - | |<partinfo>BBa_J23114</partinfo>|| || || | + | |<partinfo>BBa_J23114</partinfo> || 28.97 <br> ± <br> 0.49 || 59.44 <br> ± <br> 5.20 || 80.12 <br> ± <br> 0.95 |
|- | |- | ||
| - | |<partinfo>BBa_J23116</partinfo>|| || || | + | |<partinfo>BBa_J23116</partinfo> || 28.14 <br> ± <br> 0.25 || 72.74 <br> ± <br> 0.37 || 81.68 <br> ± <br> 3.08 |
|- | |- | ||
| - | |<partinfo>BBa_J23118</partinfo>|| || || | + | |<partinfo>BBa_J23118</partinfo> || 32.84 <br> ± <br> 0.31 || 73.64 <br> ± <br> 2.41 || 89.86 <br> ± <br> 2.93 |
|} | |} | ||
| - | '''Discussion''': we observed that the ranking previously documented in the Registry is not valid in all the conditions, even if a general agreement can be observed. As an example, <partinfo>BBa_J23110</partinfo> in high copy plasmid is stronger than <partinfo>BBa_J23118</partinfo>, in contrast with the ranking reported in the Registry. | + | '''Discussion''': we observed that the ranking previously documented in the Registry is not valid in all the tested conditions, even if a general agreement can be observed. As an example, <partinfo>BBa_J23110</partinfo> in high copy plasmid is stronger than <partinfo>BBa_J23118</partinfo>, in contrast with the ranking reported in the Registry. |
---- | ---- | ||
| Line 177: | Line 177: | ||
|} | |} | ||
| - | Some of the promoters could not be cloned upstream of these devices because they produced LuxI protein | + | Some of the promoters could not be cloned upstream of these devices because they produced amounts of LuxI protein that give a high metabolic burden for ''E. coli'', so it was not possible to study all the combinations as transformants could not be obtained in some cases. |
| - | For each part, a measurement system was built, exploiting the production of the reporter gene GFP (Green | + | For each part, a measurement system was built, exploiting the production of the reporter gene GFP (Green Fluorescent Protein) to evaluate the "switch on" condition of every self-inducible promoter. Many different combinations were explored, in order to provide a library of promoters able to initiate transcription at the desired culture density. |
| - | ===<b>Quantification of the HSL produced</b>=== | + | ===<b>Quantification of the HSL produced by autoinducer generators</b>=== |
| - | '''Experimental implementation''' The | + | '''Experimental implementation:''' The autoinducer generators <partinfo>BBa_K300030</partinfo>, <partinfo>BBa_K300028</partinfo>, <partinfo>BBa_K300029</partinfo>, <partinfo>BBa_K300025</partinfo>, <partinfo>BBa_K300026</partinfo> and <partinfo>BBa_K300027</partinfo> were, thus, characterized by measuring the concentration of HSL released in the medium of cultures grown for 6 hours. All the details are available in [[Team:UNIPV-Pavia/Parts/Characterization#Microplate reader experiments for 3OC6-HSL quantification by means of BBa_T9002 biosensor - Protocol #3|this section]]. |
<partinfo>BBa_T9002</partinfo> contained in <partinfo>pSB1A3</partinfo> in ''E. coli'' TOP10 was used as a HSL->GFP biosensor. In every experiment, a HSL-GFP calibration curve with known concentration of HSL was produced. | <partinfo>BBa_T9002</partinfo> contained in <partinfo>pSB1A3</partinfo> in ''E. coli'' TOP10 was used as a HSL->GFP biosensor. In every experiment, a HSL-GFP calibration curve with known concentration of HSL was produced. | ||
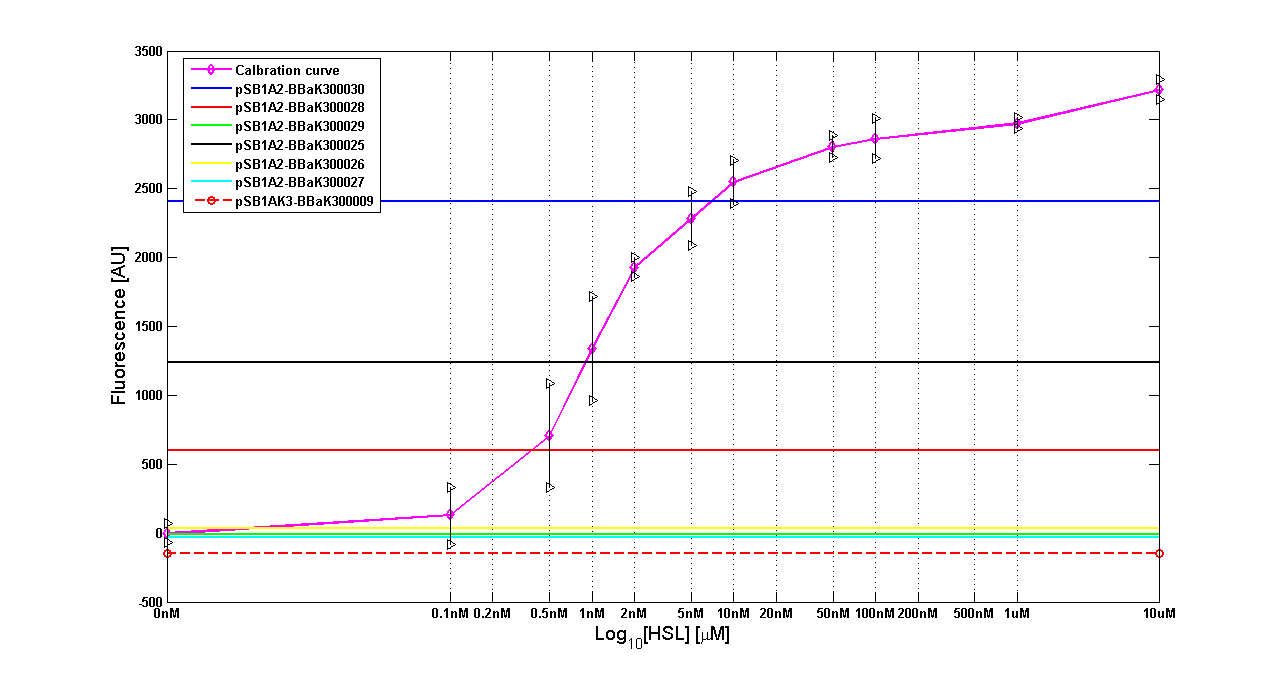
| - | '''Results''' The amount of 3OC6-HSL produced after a 6 hours growth by ''E. coli'' DH5alpha bearing the parts | + | '''Results:''' The amount of 3OC6-HSL produced after a 6 hours growth by ''E. coli'' DH5alpha bearing the parts in high copy plasmid <partinfo>pSB1A2</partinfo> is reported in Fig.8 and in the table below: |
{| align='center' | {| align='center' | ||
| - | |[[Image:pv_HCT9002sensor.png|500px|thumb|center|Figure 8 - <partinfo>BBa_T9002</partinfo> calibration curve for detection of [HSL] produced in high copy | + | |[[Image:pv_HCT9002sensor.png|500px|thumb|center|Figure 8 - <partinfo>BBa_T9002</partinfo> calibration curve for detection of [HSL] produced by autoinducer generators in high copy vector.]] |
|} | |} | ||
| Line 208: | Line 208: | ||
|} | |} | ||
| - | The amount of 3OC6-HSL produced | + | The amount of 3OC6-HSL produced by the parts contained in low copy plasmid <partinfo>pSB4C5</partinfo> after a 6 hour cell growth is reported in Fig.9 and in the table below: |
{| align='center' | {| align='center' | ||
| - | |[[Image:pv_LCT9002sensor.png|500px|thumb|center|Figure 9 - <partinfo>BBa_T9002</partinfo> calibration curve for detection of [HSL] produced in low copy | + | |[[Image:pv_LCT9002sensor.png|500px|thumb|center|Figure 9 - <partinfo>BBa_T9002</partinfo> calibration curve for detection of [HSL] produced by the autoinducer generators in low copy vector.]] |
|} | |} | ||
| Line 236: | Line 236: | ||
Signal generator and sensor device were assembled in an unique part (such as <partinfo>BBa_K300017</partinfo>, <partinfo>BBa_K300014</partinfo>, <partinfo>BBa_K300015</partinfo>, <partinfo>BBa_K300016</partinfo> and <partinfo>BBa_K300012</partinfo>) in high copy number plasmid <partinfo>pSB1A2</partinfo> or low copy number plasmid <partinfo>pSb4C5</partinfo>. A third alternative was the assembly of signal generator on a low copy number plasmid (<partinfo>pSB4C5</partinfo>) and the receiver device on high copy number plasmid (<partinfo>pSB1A2</partinfo>). | Signal generator and sensor device were assembled in an unique part (such as <partinfo>BBa_K300017</partinfo>, <partinfo>BBa_K300014</partinfo>, <partinfo>BBa_K300015</partinfo>, <partinfo>BBa_K300016</partinfo> and <partinfo>BBa_K300012</partinfo>) in high copy number plasmid <partinfo>pSB1A2</partinfo> or low copy number plasmid <partinfo>pSb4C5</partinfo>. A third alternative was the assembly of signal generator on a low copy number plasmid (<partinfo>pSB4C5</partinfo>) and the receiver device on high copy number plasmid (<partinfo>pSB1A2</partinfo>). | ||
| - | The circuits we obtained and tested are summarized | + | The circuits we obtained and tested are summarized in tables. |
| + | |||
| + | <div align='center>Sender/Receiver devices assembled as a unique BioBrick part on the same vector</div> | ||
{| border='1' align='center' | {| border='1' align='center' | ||
| - | | '''BioBrick'''<br> '''Sender''' ||'''Description ''' || '''Sender | + | | '''BioBrick'''<br> '''Sender''' ||'''Description ''' || '''Sender/Receiver Device Vector'''|| '''BioBrick composite part''' |
|- | |- | ||
| <partinfo>BBa_K300030</partinfo> | | <partinfo>BBa_K300030</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | ||
| - | + | | <partinfo>pSB1A2</partinfo><br>HC | |
|<partinfo>BBa_K300017</partinfo> | |<partinfo>BBa_K300017</partinfo> | ||
|- | |- | ||
| <partinfo>BBa_K300028</partinfo> | | <partinfo>BBa_K300028</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | ||
| - | + | | <partinfo>pSB1A2</partinfo><br>HC | |
|<partinfo>BBa_K300014</partinfo> | |<partinfo>BBa_K300014</partinfo> | ||
|- | |- | ||
| <partinfo>BBa_K300029</partinfo> | | <partinfo>BBa_K300029</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | ||
| - | + | | <partinfo>pSB1A2</partinfo><br>HC | |
|<partinfo>BBa_K300016</partinfo> | |<partinfo>BBa_K300016</partinfo> | ||
|- | |- | ||
| <partinfo>BBa_K300026</partinfo> | | <partinfo>BBa_K300026</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | ||
| - | + | | <partinfo>pSB1A2</partinfo><br>HC | |
|<partinfo>BBa_K300012</partinfo> | |<partinfo>BBa_K300012</partinfo> | ||
|- | |- | ||
| xxx|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23114 | | xxx|| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23114 | ||
| - | + | | <partinfo>pSB1A2</partinfo><br>HC | |
|<partinfo>BBa_K300015</partinfo> | |<partinfo>BBa_K300015</partinfo> | ||
|- | |- | ||
| <partinfo>BBa_K300030</partinfo> | | <partinfo>BBa_K300030</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | ||
| - | + | | <partinfo>pSB4C5</partinfo><br>LC | |
|<partinfo>BBa_K300017</partinfo> | |<partinfo>BBa_K300017</partinfo> | ||
|- | |- | ||
| <partinfo>BBa_K300028</partinfo> | | <partinfo>BBa_K300028</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | ||
| - | + | | <partinfo>pSB4C5</partinfo><br>LC | |
|<partinfo>BBa_K300014</partinfo> | |<partinfo>BBa_K300014</partinfo> | ||
|- | |- | ||
| <partinfo>BBa_K300029</partinfo> | | <partinfo>BBa_K300029</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | ||
| - | + | | <partinfo>pSB4C5</partinfo><br>LC | |
|<partinfo>BBa_K300016</partinfo> | |<partinfo>BBa_K300016</partinfo> | ||
|- | |- | ||
| <partinfo>BBa_K300026</partinfo> | | <partinfo>BBa_K300026</partinfo> | ||
| [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | | [[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23105 | ||
| - | + | | <partinfo>pSB4C5</partinfo><br>LC | |
|<partinfo>BBa_K300012</partinfo> | |<partinfo>BBa_K300012</partinfo> | ||
| + | |} | ||
| + | |||
| + | <div align='center>Sender devices assembled on low copy number vector and Receiver device on high copy number vector</div> | ||
| + | |||
| + | {| border='1' align='center' | ||
| + | | '''BioBrick'''<br> '''Sender''' ||'''Description ''' || '''Sender Vector''' || '''<partinfo>BBa_F2620</partinfo><br> Receiver vector''' | ||
|- | |- | ||
| <partinfo>BBa_K300030</partinfo> | | <partinfo>BBa_K300030</partinfo> | ||
| Line 290: | Line 298: | ||
|<partinfo>pSB4C5</partinfo><br>LC | |<partinfo>pSB4C5</partinfo><br>LC | ||
| <partinfo>pSB1A2</partinfo><br>HC | | <partinfo>pSB1A2</partinfo><br>HC | ||
| - | |||
|- | |- | ||
| <partinfo>BBa_K300028</partinfo> | | <partinfo>BBa_K300028</partinfo> | ||
| Line 296: | Line 303: | ||
|<partinfo>pSB4C5</partinfo><br>LC | |<partinfo>pSB4C5</partinfo><br>LC | ||
| <partinfo>pSB1A2</partinfo><br>HC | | <partinfo>pSB1A2</partinfo><br>HC | ||
| - | |||
|- | |- | ||
| <partinfo>BBa_K300029</partinfo> | | <partinfo>BBa_K300029</partinfo> | ||
| Line 302: | Line 308: | ||
|<partinfo>pSB4C5</partinfo><br>LC | |<partinfo>pSB4C5</partinfo><br>LC | ||
| <partinfo>pSB1A2</partinfo><br>HC | | <partinfo>pSB1A2</partinfo><br>HC | ||
| - | |||
|- | |- | ||
| <partinfo>BBa_K300025</partinfo> | | <partinfo>BBa_K300025</partinfo> | ||
| Line 308: | Line 313: | ||
|<partinfo>pSB4C5</partinfo><br>LC | |<partinfo>pSB4C5</partinfo><br>LC | ||
| <partinfo>pSB1A2</partinfo><br>HC | | <partinfo>pSB1A2</partinfo><br>HC | ||
| - | |||
|- | |- | ||
| <partinfo>BBa_K300026</partinfo> | | <partinfo>BBa_K300026</partinfo> | ||
| Line 314: | Line 318: | ||
|<partinfo>pSB4C5</partinfo><br>LC | |<partinfo>pSB4C5</partinfo><br>LC | ||
| <partinfo>pSB1A2</partinfo><br>HC | | <partinfo>pSB1A2</partinfo><br>HC | ||
| - | |||
|- | |- | ||
| <partinfo>BBa_K300027</partinfo> | | <partinfo>BBa_K300027</partinfo> | ||
| Line 320: | Line 323: | ||
|<partinfo>pSB4C5</partinfo><br>LC | |<partinfo>pSB4C5</partinfo><br>LC | ||
| <partinfo>pSB1A2</partinfo><br>HC | | <partinfo>pSB1A2</partinfo><br>HC | ||
| - | |||
|} | |} | ||
| Line 327: | Line 329: | ||
The following measurement systems were realized assembling GFP downstream of each self-inducible device. The parts characterized are reported in this table: | The following measurement systems were realized assembling GFP downstream of each self-inducible device. The parts characterized are reported in this table: | ||
| - | {| border='1' align='center' | + | {| border='1' align='center' width='80%' |
| '''Sender device''' | | '''Sender device''' | ||
| '''Sensor systems with GFP''' | | '''Sensor systems with GFP''' | ||
| Line 370: | Line 372: | ||
|<partinfo>BBa_K300030</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | |<partinfo>BBa_K300030</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23118 | ||
|<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | ||
| - | |Sender and Receiver are contained in two different plasmids, <br>cotransformed in the same cell | + | |Sender and Receiver are contained <br>in two different plasmids, <br>cotransformed in the same cell |
|- | |- | ||
|<partinfo>BBa_K300028</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | |<partinfo>BBa_K300028</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23110 | ||
|<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | |<partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo><br>[[Image:pv_T9002.png|300px]] | ||
| - | |Sender and Receiver are contained in two different plasmids, <br>cotransformed in the same cell | + | |Sender and Receiver are contained <br>in two different plasmids, <br>cotransformed in the same cell |
|- | |- | ||
|<partinfo>BBa_K300029</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | |<partinfo>BBa_K300029</partinfo> in <partinfo>pSB4C5</partinfo><br>[[Image:pv_SignalGeneratorDevice.png|150px]]<br>J23116 | ||
| Line 393: | Line 395: | ||
|} | |} | ||
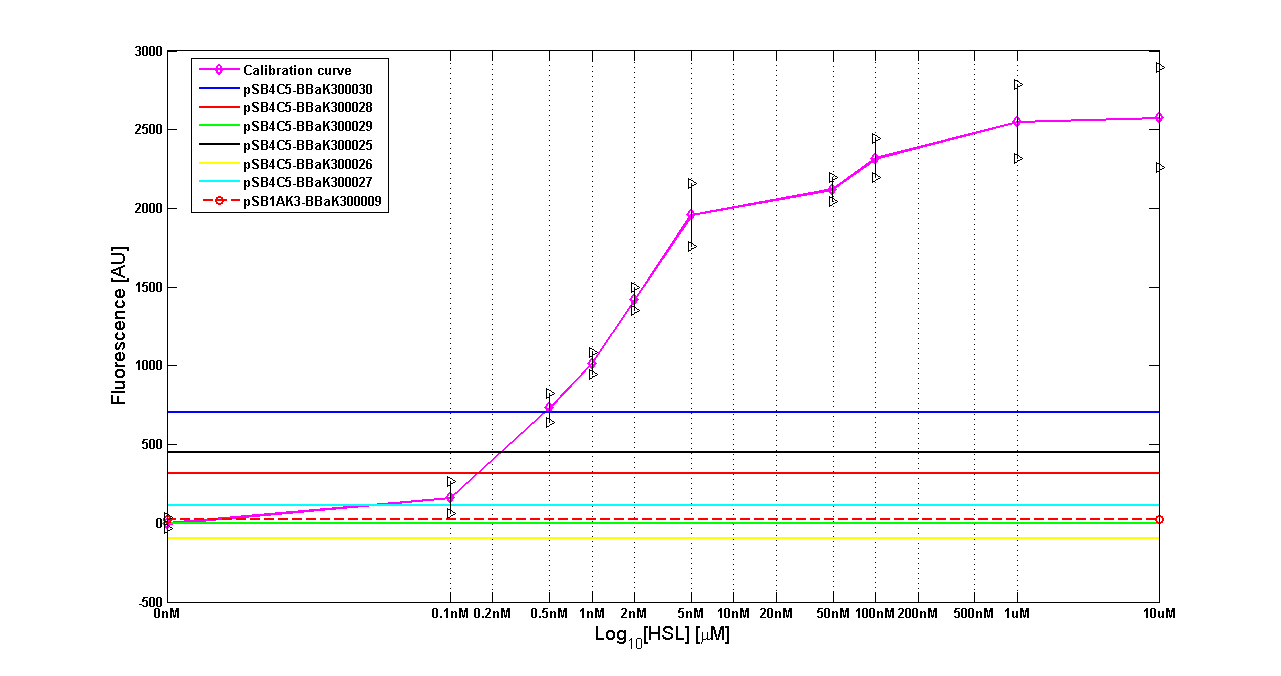
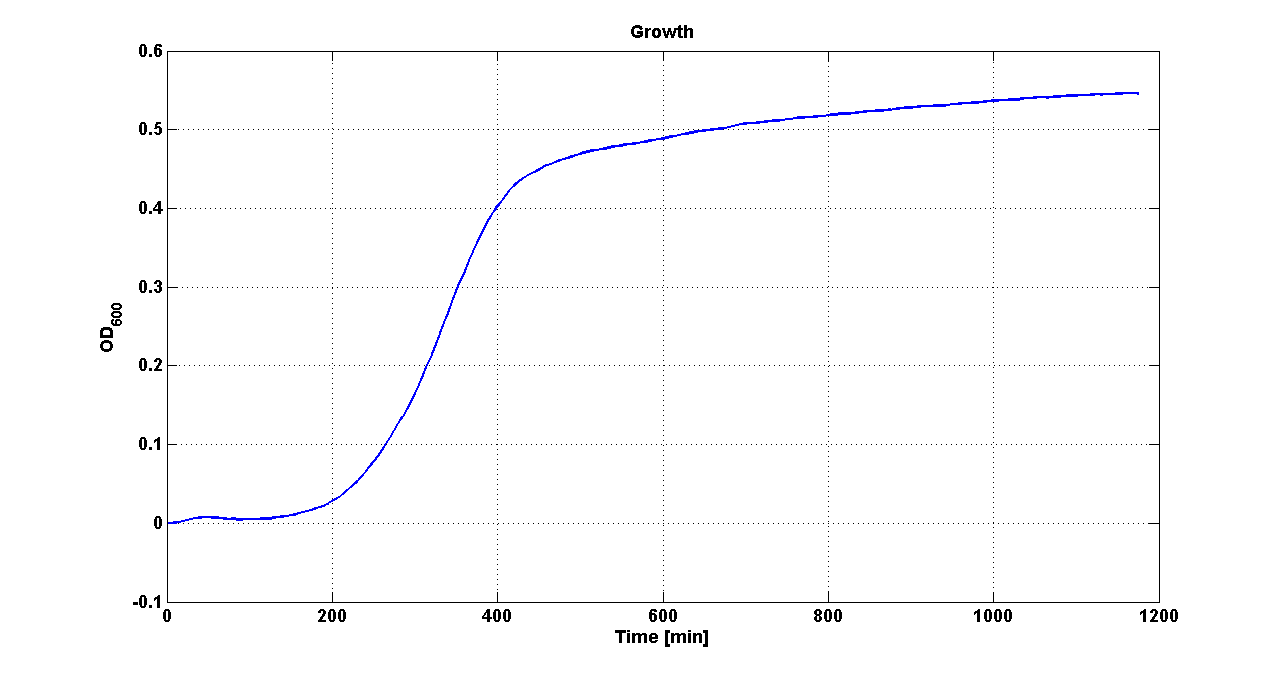
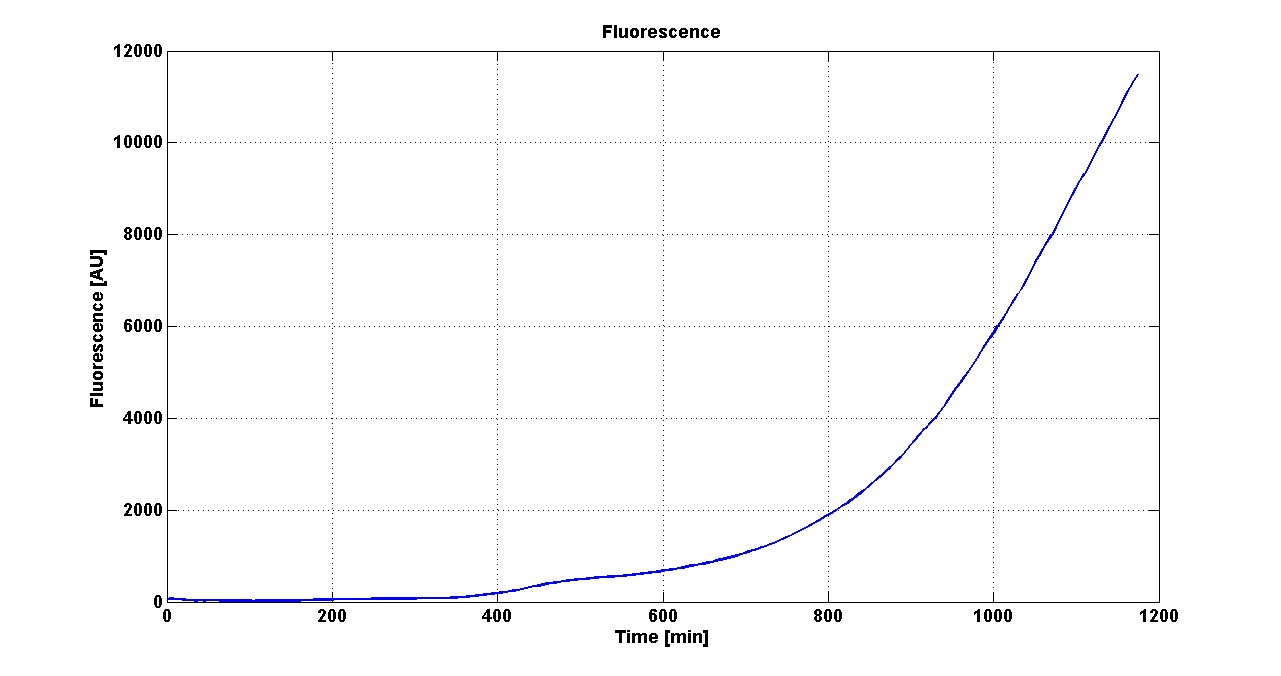
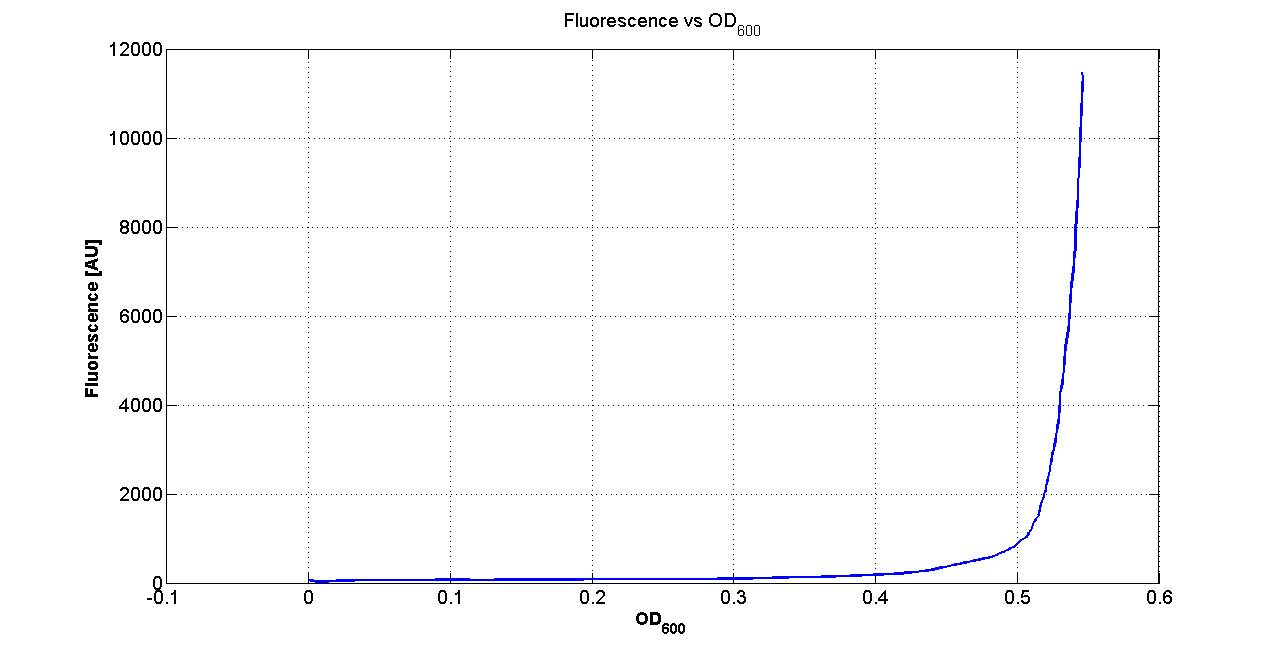
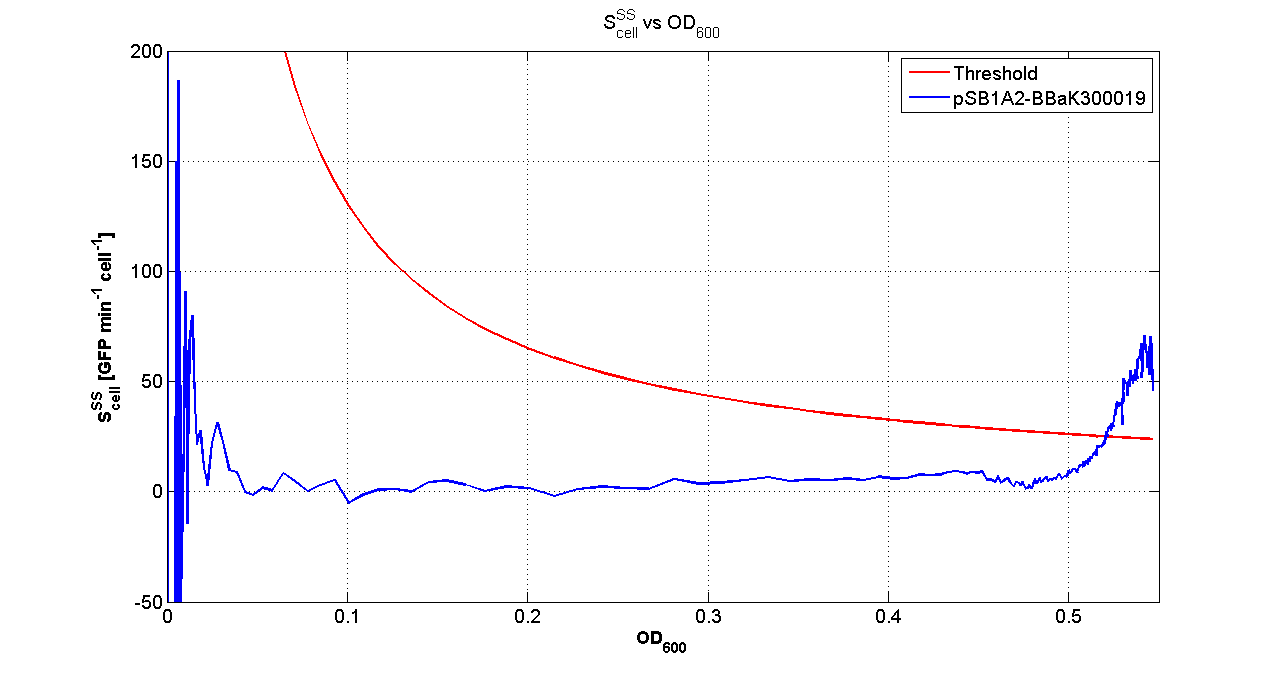
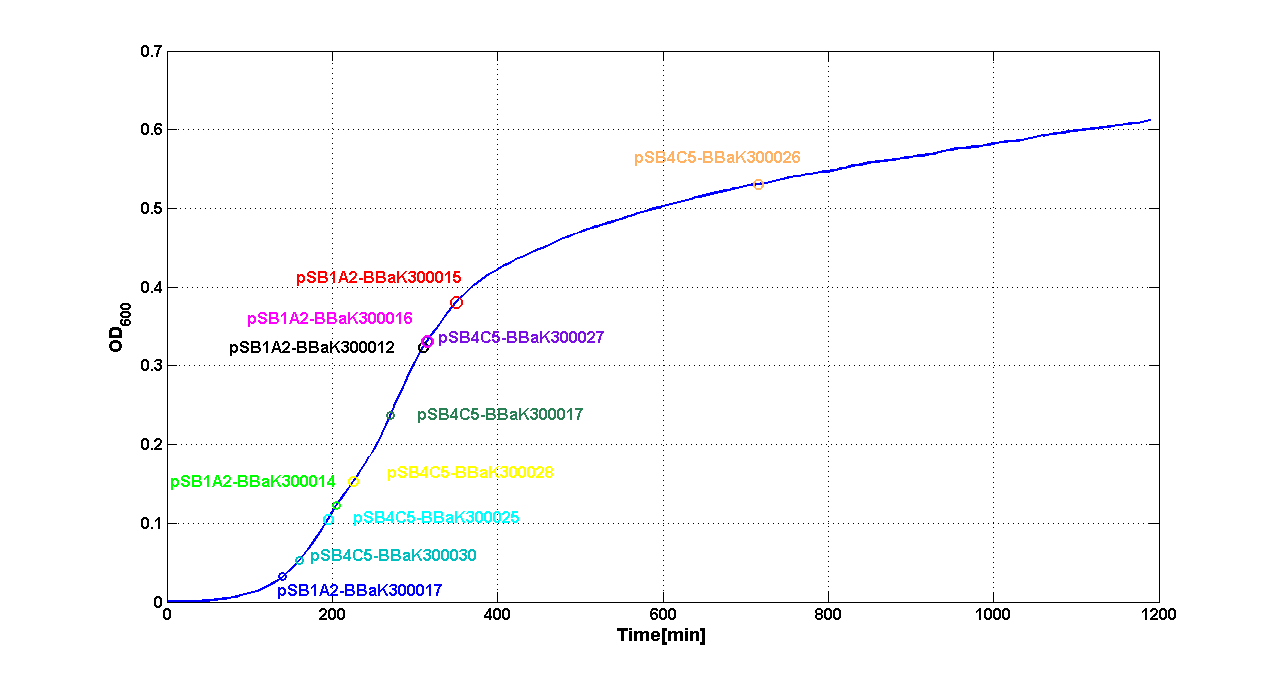
| - | Cultures of ''E. coli'' TOP10 bearing the plasmids containing the self-inducible devices expressing | + | Cultures of ''E. coli'' TOP10 bearing the plasmids containing the self-inducible devices expressing GFP were grown according to [[Team:UNIPV-Pavia/Parts/Characterization#Microplate reader experiments for self-inducible promoters - Protocol #1|this protocol]] and all data collected were analyzed as explained in [[Team:UNIPV-Pavia/Parts/Characterization#Data analysis for self-inducible promoters (initiation-treshold determination)|this section]]. An example of O.D.600 and fluorescence signals for a self-inducible device expressing GFP (<partinfo>BBa_K300026</partinfo>), as well as its Scell signal and the estimated threshold value, is reported below. |
{| | {| | ||
| - | |[[Image:pv_GrowthCurveSelf.png| | + | |[[Image:pv_GrowthCurveSelf.png|300px|thumb|center|Growth curve of <partinfo>BBa_K300019</partinfo> (O.D.600)]] |
| - | |[[Image:pv_FluoCurveSelf.png| | + | |[[Image:pv_FluoCurveSelf.png|300px|thumb|center|Fluorescence curve of <partinfo>BBa_K300019</partinfo> (G.F.P.)]] |
|- | |- | ||
| - | |[[Image:pv_FLUvsASB.png| | + | |[[Image:pv_FLUvsASB.png|300px|thumb|center|Fluorescence VS Optical density curve of <partinfo>BBa_K300019</partinfo>]] |
| - | |[[Image:pv_Scell_Threshold.png| | + | |[[Image:pv_Scell_Threshold.png|300px|thumb|center|Scell=(dGFP/dt)/O.D.600 and threshold]] |
|} | |} | ||
| - | |||
| - | |||
For every self-inducible device, several parameters were evaluated: | For every self-inducible device, several parameters were evaluated: | ||
| - | *O.D.start is the O.D.600 corresponding to the transcription initiation of the gene of interest | + | *O.D.start is the O.D.600 corresponding to the transcription initiation of the gene of interest; it was evaluated as reported [[Team:UNIPV-Pavia/Parts/Characterization#Data analysis for self-inducible promoters (initiation-treshold determination)|in this section]]; |
| - | *K_HSL is the HSL | + | *K_HSL is the HSL synthesis rate per cell; it was estimated with the algorithm described [[Team:UNIPV-Pavia/Parts/Characterization#Data analysis to estimate the HSL synthesis rate per cell|here]] |
| - | *Doubling time is the period of time required for a cell population to double | + | *Doubling time is the period of time required for a cell population to double; it was evaluated as described in [[Team:UNIPV-Pavia/Parts/Characterization#Doubling time evaluation|Doubling time evaluation section]] |
*Scell_ratio was evaluated as (Scell_max_Phi)/(Scell_max_J101). Phi is the self-inducible device of ineterst, J101 is the reference standard <partinfo>BBa_J23101</partinfo> contained in the same vector of the receiver device. Scell_max_phi was evaluated for times subsequent to the transcription initiation. | *Scell_ratio was evaluated as (Scell_max_Phi)/(Scell_max_J101). Phi is the self-inducible device of ineterst, J101 is the reference standard <partinfo>BBa_J23101</partinfo> contained in the same vector of the receiver device. Scell_max_phi was evaluated for times subsequent to the transcription initiation. | ||
Results are summarized in the following tables: | Results are summarized in the following tables: | ||
| + | |||
'''Tab. 1 - Sender and Receiver on high copy plasmid <partinfo>pSB1A2</partinfo>''' | '''Tab. 1 - Sender and Receiver on high copy plasmid <partinfo>pSB1A2</partinfo>''' | ||
| - | {| border='1' | + | {| border='1' width='80%' |
|rowspan='2' align='center'|'''Self-inducible device''' <br><font color='#50C878'>working</font><br><font color='red'>not working</font> | |rowspan='2' align='center'|'''Self-inducible device''' <br><font color='#50C878'>working</font><br><font color='red'>not working</font> | ||
| - | |||
|colspan='4' style="background: yellow" align='center' |LB | |colspan='4' style="background: yellow" align='center' |LB | ||
|colspan='4' style="background: cyan" align="center" |M9 | |colspan='4' style="background: cyan" align="center" |M9 | ||
| Line 432: | Line 432: | ||
|- | |- | ||
|<font color='#50C878'><partinfo>BBa_K300017</partinfo> (wiki name: I7) in <partinfo>pSB1A2</partinfo> plasmid</font> | |<font color='#50C878'><partinfo>BBa_K300017</partinfo> (wiki name: I7) in <partinfo>pSB1A2</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300017.png|300px]] | |
| Constitutive | | Constitutive | ||
| 3.11 10^-16 <br> ± <br> 2.23 10^-17 | | 3.11 10^-16 <br> ± <br> 2.23 10^-17 | ||
| Line 443: | Line 443: | ||
|- | |- | ||
|<font color='#50C878'><partinfo>BBa_K300014</partinfo> (wiki name: I8) in <partinfo>pSB1A2</partinfo> plasmid</font> | |<font color='#50C878'><partinfo>BBa_K300014</partinfo> (wiki name: I8) in <partinfo>pSB1A2</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300014.png|300px]] | |
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 454: | Line 454: | ||
|- | |- | ||
|<font color='#50C878'><partinfo>BBa_K300015</partinfo> (wiki name: I9) in <partinfo>pSB1A2</partinfo> plasmid</font> | |<font color='#50C878'><partinfo>BBa_K300015</partinfo> (wiki name: I9) in <partinfo>pSB1A2</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300015.png|300px]] | |
| - | | 0.33 | + | | 0.33 <br> * |
| - | | 8.94 10^-17 | + | | 8.94 10^-17 <br> * |
| 33.81 <br> ± <br> 3.03 | | 33.81 <br> ± <br> 3.03 | ||
| - | | 0.98 | + | | 0.98 <br> * |
| 0.38 <br> ± <br> 0.02 | | 0.38 <br> ± <br> 0.02 | ||
| 3.78 10^-17 <br> ± <br> 4.65 10^-18 | | 3.78 10^-17 <br> ± <br> 4.65 10^-18 | ||
| Line 465: | Line 465: | ||
|- | |- | ||
|<font color='#50C878'><partinfo>BBa_K300016</partinfo> (wiki name: I10) in <partinfo>pSB1A2</partinfo> plasmid</font> | |<font color='#50C878'><partinfo>BBa_K300016</partinfo> (wiki name: I10) in <partinfo>pSB1A2</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300016.png|300px]] | |
| - | | 0.54 | + | | 0.54 <br> * |
| - | | 7.53 10^-18 | + | | 7.53 10^-18 <br> * |
| 39.55 <br> ± <br> 0.32 | | 39.55 <br> ± <br> 0.32 | ||
| - | | 0.21 | + | | 0.21 <br> * |
| 0.32 <br> ± <br> 0.02 | | 0.32 <br> ± <br> 0.02 | ||
| 5.70 10^-17 <br> ± <br> 7.75 10^-18 | | 5.70 10^-17 <br> ± <br> 7.75 10^-18 | ||
| Line 476: | Line 476: | ||
|- | |- | ||
|<font color='#50C878'><partinfo>BBa_K300012</partinfo> (wiki name: I12) in <partinfo>pSB1A2</partinfo> plasmid</font> | |<font color='#50C878'><partinfo>BBa_K300012</partinfo> (wiki name: I12) in <partinfo>pSB1A2</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300012.png|300px]] | |
| 0.50 <br> ± <br> 0.01 | | 0.50 <br> ± <br> 0.01 | ||
| 1.16 10^-17 <br> ± <br> 6.41 10^-19 | | 1.16 10^-17 <br> ± <br> 6.41 10^-19 | ||
| Line 486: | Line 486: | ||
| 0.21 <br> ± <br> 0.06 | | 0.21 <br> ± <br> 0.06 | ||
|} | |} | ||
| + | |||
| + | |||
'''Tab. 2 - Sender and Receiver on low copy plasmid <partinfo>pSB4C5</partinfo>''' | '''Tab. 2 - Sender and Receiver on low copy plasmid <partinfo>pSB4C5</partinfo>''' | ||
| - | {| border='1' | + | {| border='1' width='80%' |
|rowspan='2' align='center'|'''Self-inducible device''' <br><font color='#50C878'>working</font><br><font color='red'>not working</font> | |rowspan='2' align='center'|'''Self-inducible device''' <br><font color='#50C878'>working</font><br><font color='red'>not working</font> | ||
| - | |||
|colspan='4' style="background: yellow" align='center' |LB | |colspan='4' style="background: yellow" align='center' |LB | ||
|colspan='4' style="background: cyan" align="center" |M9 | |colspan='4' style="background: cyan" align="center" |M9 | ||
| Line 504: | Line 505: | ||
|align='center'|'''Scell ratio''' | |align='center'|'''Scell ratio''' | ||
|- | |- | ||
| - | |<font color='#50C878'><partinfo>BBa_K300017</partinfo> (wiki name: I7) in <partinfo>pSB4C5</partinfo> plasmid</font> | + | |<font color='#50C878'><partinfo>BBa_K300017</partinfo> (wiki name: I7) in <partinfo>pSB4C5</partinfo> plasmid</font><br>[[Image:pv_BBa_K300017.png|300px]] |
| - | + | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 516: | Line 516: | ||
|- | |- | ||
|<font color='red'><partinfo>BBa_K300014</partinfo> (wiki name: I8) in <partinfo>pSB4C5</partinfo> plasmid</font> | |<font color='red'><partinfo>BBa_K300014</partinfo> (wiki name: I8) in <partinfo>pSB4C5</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300014.png|300px]] | |
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 527: | Line 527: | ||
|- | |- | ||
|<font color='red'><partinfo>BBa_K300015</partinfo> (wiki name: I9) in <partinfo>pSB4C5</partinfo> plasmid</font> | |<font color='red'><partinfo>BBa_K300015</partinfo> (wiki name: I9) in <partinfo>pSB4C5</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300015.png|300px]] | |
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 538: | Line 538: | ||
|- | |- | ||
|<font color='red'><partinfo>BBa_K300016</partinfo> (wiki name: I10) in <partinfo>pSB4C5</partinfo> plasmid</font> | |<font color='red'><partinfo>BBa_K300016</partinfo> (wiki name: I10) in <partinfo>pSB4C5</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300016.png|300px]] | |
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 549: | Line 549: | ||
|- | |- | ||
|<font color='red'><partinfo>BBa_K300012</partinfo> (wiki name: I12) in <partinfo>pSB4C5</partinfo> plasmid</font> | |<font color='red'><partinfo>BBa_K300012</partinfo> (wiki name: I12) in <partinfo>pSB4C5</partinfo> plasmid</font> | ||
| - | + | <br>[[Image:pv_BBa_K300012.png|300px]] | |
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 559: | Line 559: | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
|} | |} | ||
| + | |||
'''Tab. 3 - Sender on low copy plasmid <partinfo>pSB4C5</partinfo> and Receiver on high copy plasmid <partinfo>pSB1A3</partinfo>''' | '''Tab. 3 - Sender on low copy plasmid <partinfo>pSB4C5</partinfo> and Receiver on high copy plasmid <partinfo>pSB1A3</partinfo>''' | ||
| - | {| border='1' | + | {| border='1' width='80%' |
|rowspan='2' align='center'|'''Self-inducible device''' <br><font color='#50C878'>working</font><br><font color='red'>not working</font> | |rowspan='2' align='center'|'''Self-inducible device''' <br><font color='#50C878'>working</font><br><font color='red'>not working</font> | ||
| - | |||
|colspan='4' style="background: yellow" align='center' |LB | |colspan='4' style="background: yellow" align='center' |LB | ||
|colspan='4' style="background: cyan" align="center" |M9 | |colspan='4' style="background: cyan" align="center" |M9 | ||
| Line 577: | Line 577: | ||
|align='center'|'''Scell ratio''' | |align='center'|'''Scell ratio''' | ||
|- | |- | ||
| - | |<font color='#50C878'><partinfo>BBa_K300030</partinfo><br> (wiki name: I14) <br>in <partinfo>pSB4C5</partinfo> plasmid</font> | + | |<table><tr><td><font color='#50C878'><partinfo>BBa_K300030</partinfo><br> (wiki name: I14) <br>in <partinfo>pSB4C5</partinfo> plasmid</font> |
| - | + | <br></td><td><partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo></td></tr> | |
| - | + | <tr><td>[[Image:pv_BBa_K300030.png|170px]]<br>LC</td> | |
| - | | Constitutive | + | <td>[[Image:pv_BBa_F2620.png|170px]]<br>HC</td></tr></table> |
| + | |Constitutive | ||
| 2.92 10^-16 <br> ± <br> 8.16 10^-18 | | 2.92 10^-16 <br> ± <br> 8.16 10^-18 | ||
| 33.20 <br> ± <br> 1.46 | | 33.20 <br> ± <br> 1.46 | ||
| Line 589: | Line 590: | ||
| 0.91 <br> ± <br> 0.15 | | 0.91 <br> ± <br> 0.15 | ||
|- | |- | ||
| - | |<font color='#50C878'><partinfo>BBa_K300028</partinfo><br> (wiki name: I15)<br> in <partinfo>pSB4C5</partinfo> plasmid</font> | + | |<table><tr><td><font color='#50C878'><partinfo>BBa_K300028</partinfo><br> (wiki name: I15)<br> in <partinfo>pSB4C5</partinfo> plasmid</font><br></td><td><partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo></td></tr> |
| - | + | <tr><td>[[Image:pv_BBa_K300028.png|170px]]<br>LC</td> | |
| - | + | <td>[[Image:pv_BBa_F2620.png|170px]]<br>HC</td></tr></table> | |
| 0.32 <br> ± <br> 0.04 ** | | 0.32 <br> ± <br> 0.04 ** | ||
| 8.31 10^-17 <br> ± <br> 3.92 10^-17 ** | | 8.31 10^-17 <br> ± <br> 3.92 10^-17 ** | ||
| Line 601: | Line 602: | ||
| 0.26 <br> ± <br> 0.03 | | 0.26 <br> ± <br> 0.03 | ||
|- | |- | ||
| - | |<font color='#50C878'><partinfo>BBa_K300029</partinfo | + | |<table><tr><td><font color='#50C878'><partinfo>BBa_K300029</partinfo> (wiki name: I16) in <partinfo>pSB4C5</partinfo> plasmid</font> |
| - | + | </td><td><partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo></td></tr> | |
| - | + | <tr><td>[[Image:pv_BBa_K300029.png|170px]]<br>LC</td> | |
| - | | 0.31 | + | <td>[[Image:pv_BBa_F2620.png|170px]]<br>HC</td></tr></table> |
| - | | 1.17 10^-16 | + | | 0.31 <br> * |
| + | | 1.17 10^-16 <br> * | ||
| 35.46 <br> ± <br> 2.85 | | 35.46 <br> ± <br> 2.85 | ||
| - | | 0.57 | + | | 0.57 <br> * |
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 613: | Line 615: | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
|- | |- | ||
| - | |<font color='#50C878'><partinfo>BBa_K300025</partinfo><br> (wiki name: I17) <br>in <partinfo>pSB4C5</partinfo> plasmid</font> | + | |<table><tr><td><font color='#50C878'><partinfo>BBa_K300025</partinfo><br> (wiki name: I17) <br>in <partinfo>pSB4C5</partinfo> plasmid</font> |
| - | + | </td><td><partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo></td></tr> | |
| - | + | <tr><td>[[Image:pv_BBa_K300025.png|170px]]<br>LC</td> | |
| + | <td>[[Image:pv_BBa_F2620.png|170px]]<br>HC</td></tr></table> | ||
| Constitutive | | Constitutive | ||
| 2.85 10^-16 <br> ± <br> 1.90 10^-17 | | 2.85 10^-16 <br> ± <br> 1.90 10^-17 | ||
| Line 625: | Line 628: | ||
| 0.45 <br> ± <br> 0.08 | | 0.45 <br> ± <br> 0.08 | ||
|- | |- | ||
| - | |<font color='#50C878'><partinfo>BBa_K300026</partinfo><br> (wiki name: I18) <br>in <partinfo>pSB4C5</partinfo> plasmid</font> | + | |<table><tr><td><font color='#50C878'><partinfo>BBa_K300026</partinfo><br> (wiki name: I18) <br>in <partinfo>pSB4C5</partinfo> plasmid</font> |
| - | + | </td><td><partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo></td></tr> | |
| - | + | <tr><td>[[Image:pv_BBa_K300026.png|170px]]<br>LC</td> | |
| + | <td>[[Image:pv_BBa_F2620.png|170px]]<br>HC</td></tr></table> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 637: | Line 641: | ||
| 0.03 <br> ± <br> 0.002 ** | | 0.03 <br> ± <br> 0.002 ** | ||
|- | |- | ||
| - | |<font color='#50C878'><partinfo>BBa_K300027</partinfo><br> (wiki name: I19)<br> in <partinfo>pSB4C5</partinfo> plasmid</font> | + | |<table><tr><td><font color='#50C878'><partinfo>BBa_K300027</partinfo><br> (wiki name: I19)<br> in <partinfo>pSB4C5</partinfo> plasmid</font> |
| - | + | </td><td><partinfo>BBa_T9002</partinfo> in <partinfo>pSB1A3</partinfo></td></tr> | |
| - | + | <tr><td>[[Image:pv_BBa_K300027.png|170px]]<br>LC</td> | |
| + | <td>[[Image:pv_BBa_F2620.png|170px]]<br>HC</td></tr></table> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| <font color='red' size='+2'>X</font> | | <font color='red' size='+2'>X</font> | ||
| Line 650: | Line 655: | ||
|} | |} | ||
| - | <div id="box" style="width: | + | <div id="box" style="width: 70%; margin-left: 137px; padding: 5px; border: 1px solid #000; background-color: #f6f6f6;"> |
<div id="template" style="text-align: center; font-weight: bold; font-size: large; color: black; padding: 5px;"> | <div id="template" style="text-align: center; font-weight: bold; font-size: large; color: black; padding: 5px;"> | ||
LEGEND OF TABLES: | LEGEND OF TABLES: | ||
</div> | </div> | ||
<div id="legend" style="font-weight: normal; font-size: small; color: black; padding: 5px;"> | <div id="legend" style="font-weight: normal; font-size: small; color: black; padding: 5px;"> | ||
| - | ''Constitutive'': the induction point, in term of O.D.600, is under the minimum detectable value calculated by the | + | ''Constitutive'': the induction point, in term of O.D.600, is under the minimum detectable value calculated by the algorithm. This minimum value was estimated by running the algorithm on data acquired from a culture that constitutively produces GFP. For this reason, the devices labelled as ''constitutive'' can be considered as constitutive GFP producers. |
<nowiki>*</nowiki>: in two of three experiments the self-induction failed, thus having a non-induced culture for all the cell densities. The standard errors were not computed for these cultures. | <nowiki>*</nowiki>: in two of three experiments the self-induction failed, thus having a non-induced culture for all the cell densities. The standard errors were not computed for these cultures. | ||
| - | <nowiki>**</nowiki>: in one of three experiments the self-induction failed, thus having a non-induced culture for all the cell densities. The standard errors were | + | <nowiki>**</nowiki>: in one of three experiments the self-induction failed, thus having a non-induced culture for all the cell densities. The standard errors were computed on two independent experiments. |
| - | <partinfo>BBa_K300016</partinfo> is labelled with <nowiki>*</nowiki>, but probably induction failed in two of the three experiments because the culture didn't reach the | + | <partinfo>BBa_K300016</partinfo> is labelled with <nowiki>*</nowiki>, but probably induction failed in two of the three experiments because the culture didn't reach the O.D.start point (the experiment was stopped before the culture reached the O.D.600 critical value). |
</div> | </div> | ||
</div> | </div> | ||
| - | '''Discussion''': two | + | '''Discussion''': two modular PoPS-based devices (<partinfo>BBa_K300010</partinfo> and <partinfo>BBa_K300009</partinfo>/<partinfo>BBa_F2620</partinfo>) were designed and used to realize a library of self-inducible devices, able to start the production of the heterologous protein at a defined culture density. They were characterized in many different experimental conditions: |
*varying the strength of the promoter controlling the production of the signal molecule (Sender Modulation) | *varying the strength of the promoter controlling the production of the signal molecule (Sender Modulation) | ||
*varying the copy number of vectors bearing both Sender and Receiver circuits | *varying the copy number of vectors bearing both Sender and Receiver circuits | ||
*varying the growth medium (LB or M9) | *varying the growth medium (LB or M9) | ||
| - | + | A graphical summary is reported in the figures below: | |
{| | {| | ||
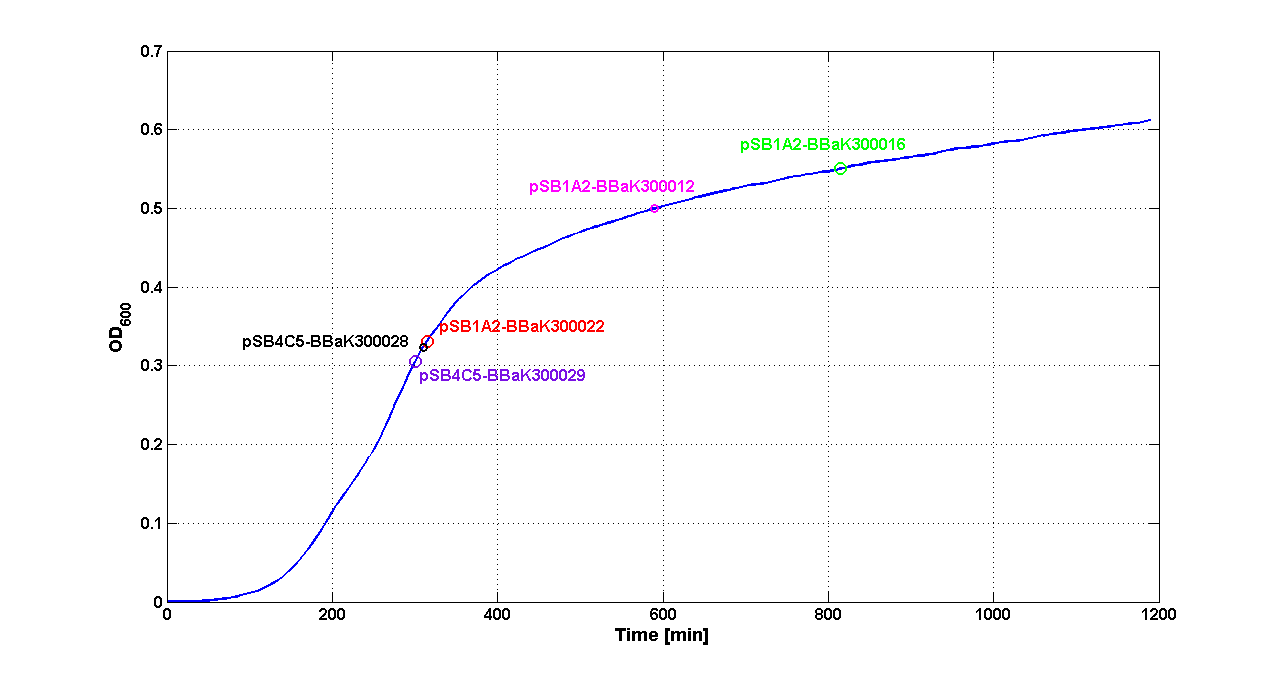
| - | |[[Image:pv_SwitchPointLB.png|700px|thumb|center| | + | |[[Image:pv_SwitchPointLB.png|700px|thumb|center| Typical ''E. coli'' growth curve with O.D.start evaluated by Threshold algorithm in LB]] |
|- | |- | ||
| - | |[[Image:pv_SwitchPointM9.png|700px|thumb|center| | + | |[[Image:pv_SwitchPointM9.png|700px|thumb|center| Typical ''E. coli'' growth curve with O.D.start evaluated by Threshold algorithm in M9]] |
|} | |} | ||
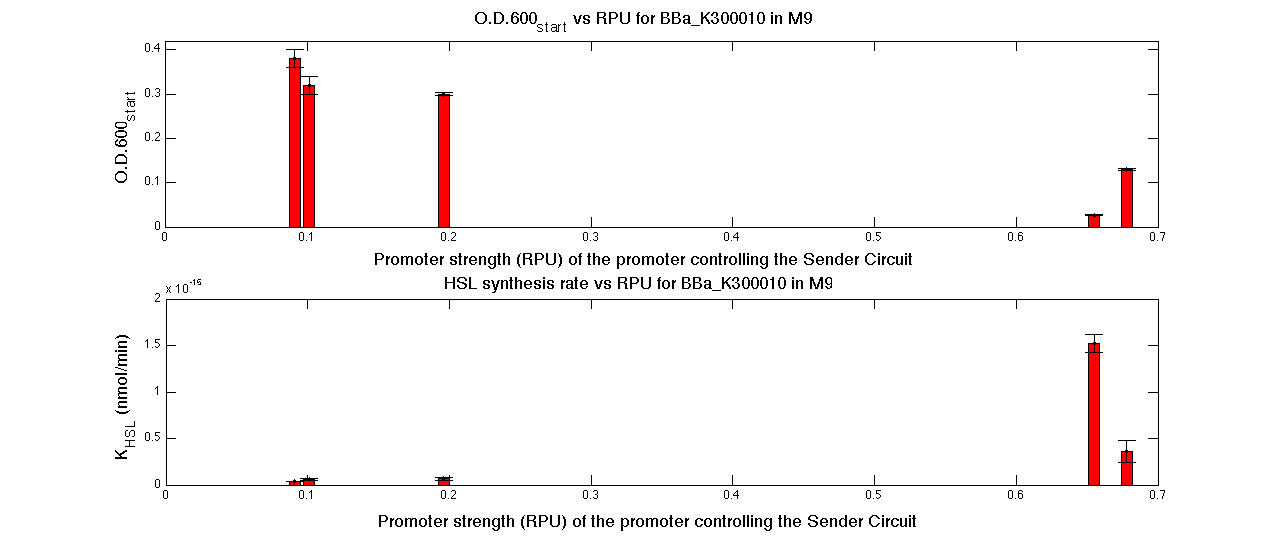
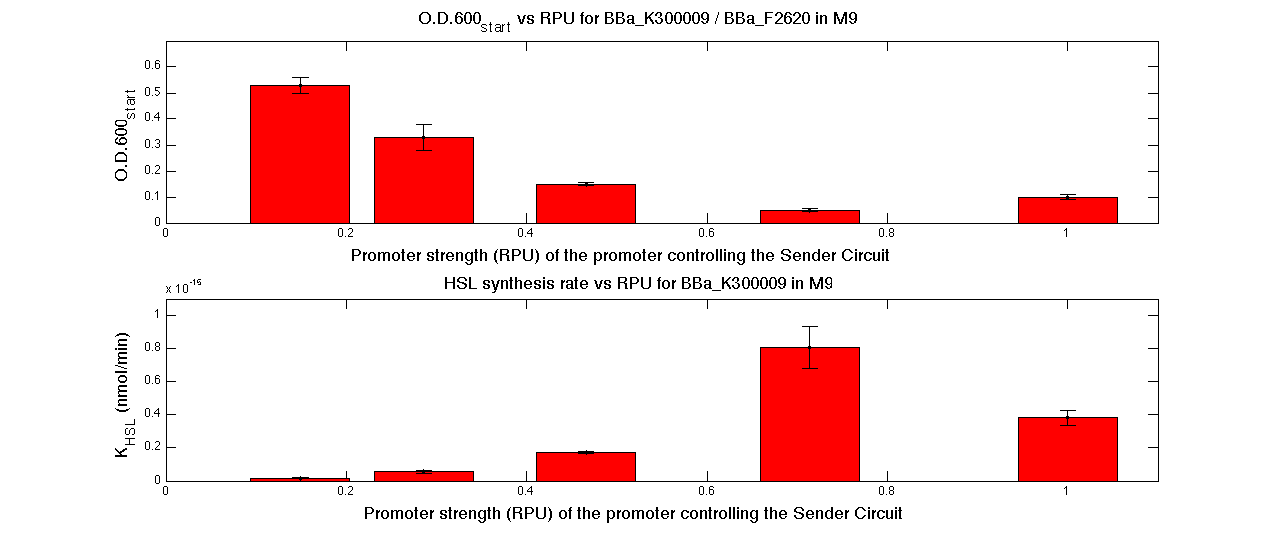
| - | A model-based approach was proposed to estimate many interesting parameters, such as the HSL synthesis rate per cell and an algorithm was proposed | + | A model-based approach was proposed to estimate many interesting parameters, such as the HSL synthesis rate per cell, and an algorithm was proposed in order to evaluate the O.D.start for every self-inducible device. |
| - | In | + | In the figures below, the O.D.start and the HSL synthesis rate as a function of the strength of the promoter (RPU) controlling the signal molecule production are reported for <partinfo>BBa_K300010</partinfo> (Sender&Receiver device in HC plasmid) and for <partinfo>BBa_K300009</partinfo>/<partinfo>BBa_F2620</partinfo> (Sender device in LC plasmid in combination with Receiver device in HC plasmid). |
{| | {| | ||
|[[Image:pv_RPUvsOD_HCHC_M9.png|700px|thumb|center| O.D.start and K_HSL as a function of RPUs of the promoters controlling the signal molecule production for <partinfo>BBa_K300010</partinfo> in high copy number plasmid in M9 medium]] | |[[Image:pv_RPUvsOD_HCHC_M9.png|700px|thumb|center| O.D.start and K_HSL as a function of RPUs of the promoters controlling the signal molecule production for <partinfo>BBa_K300010</partinfo> in high copy number plasmid in M9 medium]] | ||
|- | |- | ||
| - | |[[Image:pv_RPUvsOD_HCLC_M9.png|700px|thumb|center| O.D.start and K_HSL as a function of RPUs of the promoters controlling the | + | |[[Image:pv_RPUvsOD_HCLC_M9.png|700px|thumb|center| O.D.start and K_HSL as a function of RPUs of the promoters controlling the autoinducer production for <partinfo>BBa_K300010</partinfo> in low copy number plasmid used in combination with <partinfo>BBa_F2620</partinfo> in high copy plasmid in M9 medium]] |
|} | |} | ||
| - | A strong correlation between the promoter strength, previously measured in RPU, and the O.D.start is depicted by | + | A strong correlation between the promoter strength, previously measured in RPU, and the O.D.start is depicted by the reported graphs. This is consistent with the expected behaviour of these parts, since the HSL synthesis rate is an increasing function of the upstream promoter's strength. |
| + | |||
| + | For RPU values greater or equal to 1, the expected behaviour is not confirmed anymore. This is probably because too high luxI expression levels and/or synthesis rate of HSL are injurious for the cell. | ||
| - | + | The combination of promoter strength variation and plasmid copy number modulation allows the creation of a library of self-inducible devices able to perform autoinduction at many different O.D.600 values, in any cellular growth phase. | |
| - | + | ||
| - | The combination of promoter strength variation and plasmid copy number modulation allows the creation of a library of self-inducible devices | + | |
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
Latest revision as of 21:41, 27 October 2010
|
Self-inducible promotersRegulation of signal protein productionExperimental implementation: <partinfo>BBa_K300009</partinfo> part was assembled downstream of different constitutive promoters, thus obtaining a signal molecule generator. The choice of constitutive promoters was performed between the ones belonging to the [http://partsregistry.org/Part:BBa_J23101 Anderson’s promoters collection]; we chose promoters according to their activities reported in the Registry of Standard Biological Parts, in order to have a thick mesh:
Before constructing the signal generators, <partinfo>BBa_K300009</partinfo> and <partinfo>BBa_K300010</partinfo> under the regulation of one of these constitutive promoters, we evaluated the promoter activities in Relative Promoter Units (R.P.U.) according to Data analysis for RPU evaluation, using the reporter protein RFP (Red Fluorescent Protein) in different experimental conditions (plasmids’ copy number and growth medium), many of them not yet explored and documented:
It was not possible to evaluate promoters activities in low copy number plasmids in LB because the RFP activity was too weak and not distinguishable from the background. RFP fluorescence and Optical Density at 600nm (O.D.600) were measured in 96-well microplates, as reported in Microplate reader experiments for constitutive promoters (R.P.U. evaluation) - Protocol #2. Data were analyzed as reported in Data analysis for RPU evaluation; Results: results are shown here. Doubling times were evaluated for the described cultures (HC stands for High Copy and LC stands for Low Copy):
Discussion: we observed that the ranking previously documented in the Registry is not valid in all the tested conditions, even if a general agreement can be observed. As an example, <partinfo>BBa_J23110</partinfo> in high copy plasmid is stronger than <partinfo>BBa_J23118</partinfo>, in contrast with the ranking reported in the Registry. After the evaluation of promoter activity, signal generators were constructed in high copy and low copy plasmids: <partinfo>BBa_K300009</partinfo> and <partinfo>BBa_K300010</partinfo> were assembled downstream of the above mentioned promoters, thus obtaining the following parts: Some of the promoters could not be cloned upstream of these devices because they produced amounts of LuxI protein that give a high metabolic burden for E. coli, so it was not possible to study all the combinations as transformants could not be obtained in some cases. For each part, a measurement system was built, exploiting the production of the reporter gene GFP (Green Fluorescent Protein) to evaluate the "switch on" condition of every self-inducible promoter. Many different combinations were explored, in order to provide a library of promoters able to initiate transcription at the desired culture density. Quantification of the HSL produced by autoinducer generatorsExperimental implementation: The autoinducer generators <partinfo>BBa_K300030</partinfo>, <partinfo>BBa_K300028</partinfo>, <partinfo>BBa_K300029</partinfo>, <partinfo>BBa_K300025</partinfo>, <partinfo>BBa_K300026</partinfo> and <partinfo>BBa_K300027</partinfo> were, thus, characterized by measuring the concentration of HSL released in the medium of cultures grown for 6 hours. All the details are available in this section. <partinfo>BBa_T9002</partinfo> contained in <partinfo>pSB1A3</partinfo> in E. coli TOP10 was used as a HSL->GFP biosensor. In every experiment, a HSL-GFP calibration curve with known concentration of HSL was produced. Results: The amount of 3OC6-HSL produced after a 6 hours growth by E. coli DH5alpha bearing the parts in high copy plasmid <partinfo>pSB1A2</partinfo> is reported in Fig.8 and in the table below:
The amount of 3OC6-HSL produced by the parts contained in low copy plasmid <partinfo>pSB4C5</partinfo> after a 6 hour cell growth is reported in Fig.9 and in the table below:
Discussion These experiments provided extremely useful informations about the capability of the signal generators to produce the 3OC6-HSL signal molecule. Data are quantitative, but incomplete because for weak promoters or medium-strength promoters contained in a low copy number plasmid the amount of 3OC6-HSL was not detectable using this system. However, this simple experiment shows that there is a strong correlation between the strength of promoter and the amount of signal molecule produced. These results confirm that the production of the autoinducer can be engineered in E. coli and different expression systems reach different amounts of 3OC6-HSL in the growth media as a function of the promoter strength. Thus, these results demonstrate that self-inducible circuits can be rationally designed from a set of well characterized standard parts. Modulation of plasmid copy numberSignal generator and sensor device were assembled in an unique part (such as <partinfo>BBa_K300017</partinfo>, <partinfo>BBa_K300014</partinfo>, <partinfo>BBa_K300015</partinfo>, <partinfo>BBa_K300016</partinfo> and <partinfo>BBa_K300012</partinfo>) in high copy number plasmid <partinfo>pSB1A2</partinfo> or low copy number plasmid <partinfo>pSb4C5</partinfo>. A third alternative was the assembly of signal generator on a low copy number plasmid (<partinfo>pSB4C5</partinfo>) and the receiver device on high copy number plasmid (<partinfo>pSB1A2</partinfo>). The circuits we obtained and tested are summarized in tables.
Sender/Receiver devices assembled as a unique BioBrick part on the same vector
Sender devices assembled on low copy number vector and Receiver device on high copy number vector
ResultsThe following measurement systems were realized assembling GFP downstream of each self-inducible device. The parts characterized are reported in this table: Cultures of E. coli TOP10 bearing the plasmids containing the self-inducible devices expressing GFP were grown according to this protocol and all data collected were analyzed as explained in this section. An example of O.D.600 and fluorescence signals for a self-inducible device expressing GFP (<partinfo>BBa_K300026</partinfo>), as well as its Scell signal and the estimated threshold value, is reported below. For every self-inducible device, several parameters were evaluated:
Results are summarized in the following tables:
Tab. 1 - Sender and Receiver on high copy plasmid <partinfo>pSB1A2</partinfo>
Tab. 2 - Sender and Receiver on low copy plasmid <partinfo>pSB4C5</partinfo>
LEGEND OF TABLES: Constitutive: the induction point, in term of O.D.600, is under the minimum detectable value calculated by the algorithm. This minimum value was estimated by running the algorithm on data acquired from a culture that constitutively produces GFP. For this reason, the devices labelled as constitutive can be considered as constitutive GFP producers. *: in two of three experiments the self-induction failed, thus having a non-induced culture for all the cell densities. The standard errors were not computed for these cultures. **: in one of three experiments the self-induction failed, thus having a non-induced culture for all the cell densities. The standard errors were computed on two independent experiments. <partinfo>BBa_K300016</partinfo> is labelled with *, but probably induction failed in two of the three experiments because the culture didn't reach the O.D.start point (the experiment was stopped before the culture reached the O.D.600 critical value). Discussion: two modular PoPS-based devices (<partinfo>BBa_K300010</partinfo> and <partinfo>BBa_K300009</partinfo>/<partinfo>BBa_F2620</partinfo>) were designed and used to realize a library of self-inducible devices, able to start the production of the heterologous protein at a defined culture density. They were characterized in many different experimental conditions:
A graphical summary is reported in the figures below: A model-based approach was proposed to estimate many interesting parameters, such as the HSL synthesis rate per cell, and an algorithm was proposed in order to evaluate the O.D.start for every self-inducible device. In the figures below, the O.D.start and the HSL synthesis rate as a function of the strength of the promoter (RPU) controlling the signal molecule production are reported for <partinfo>BBa_K300010</partinfo> (Sender&Receiver device in HC plasmid) and for <partinfo>BBa_K300009</partinfo>/<partinfo>BBa_F2620</partinfo> (Sender device in LC plasmid in combination with Receiver device in HC plasmid). A strong correlation between the promoter strength, previously measured in RPU, and the O.D.start is depicted by the reported graphs. This is consistent with the expected behaviour of these parts, since the HSL synthesis rate is an increasing function of the upstream promoter's strength. For RPU values greater or equal to 1, the expected behaviour is not confirmed anymore. This is probably because too high luxI expression levels and/or synthesis rate of HSL are injurious for the cell. The combination of promoter strength variation and plasmid copy number modulation allows the creation of a library of self-inducible devices able to perform autoinduction at many different O.D.600 values, in any cellular growth phase.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"