Team:HKU-Hong Kong/Project
From 2010.igem.org
(→Results) |
(→Scenarios) |
||
| (5 intermediate revisions not shown) | |||
| Line 52: | Line 52: | ||
| - | + | *below is another gene we found it possible to achieve the cell-lysis | |
| - | + | ||
| Line 60: | Line 59: | ||
[[Image:Gene E location.JPG|500px]] | [[Image:Gene E location.JPG|500px]] | ||
| - | The figure shows the location of gene E in the phiX174 genome | + | The figure shows the location of gene ''E'' in the phiX174 genome |
(YOUNG, MICROBIOLOGIcAL REVIEWS, Sept. 1992,Vol. 56, No. 3) | (YOUNG, MICROBIOLOGIcAL REVIEWS, Sept. 1992,Vol. 56, No. 3) | ||
| Line 106: | Line 105: | ||
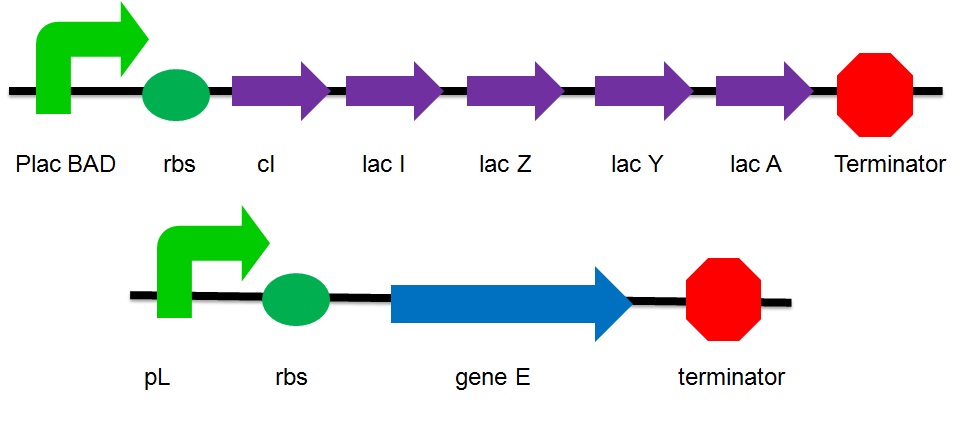
== '''The plasmid design''' == | == '''The plasmid design''' == | ||
[[Image:gene E and lac operon.JPG|700px]] | [[Image:gene E and lac operon.JPG|700px]] | ||
| - | ;PlacBAD | + | ;''PlacBAD'' |
: It is the promoter of the lac operon inducible by lactose or arabinose. Without the substrate, the promoter is suppressed by the repressor protein. Under this condition, the gene is considered as turned off. No expression of genes that are upstream the suppressed promoter can be observed. No proteins that are encoded by these genes are produced. | : It is the promoter of the lac operon inducible by lactose or arabinose. Without the substrate, the promoter is suppressed by the repressor protein. Under this condition, the gene is considered as turned off. No expression of genes that are upstream the suppressed promoter can be observed. No proteins that are encoded by these genes are produced. | ||
| - | ;cI | + | ;''cI'' |
: It is the coding sequence of cI protein. The cI protein acts as a repressor protein and bind to the promoter of the suicide gene. The suicide mechanism is then turned off without expression of gene ''E'' and production of E protein. | : It is the coding sequence of cI protein. The cI protein acts as a repressor protein and bind to the promoter of the suicide gene. The suicide mechanism is then turned off without expression of gene ''E'' and production of E protein. | ||
| - | ;Lac I | + | ;''Lac I'' |
: Encode for the inhibitor of the lac operon | : Encode for the inhibitor of the lac operon | ||
| - | ;Lac Z | + | ;''Lac Z'' |
: Encode for Beta-galactosidase which breakdowns lactose to glucose & galactose and convert lactose to L-lactose which can activate the lac operon | : Encode for Beta-galactosidase which breakdowns lactose to glucose & galactose and convert lactose to L-lactose which can activate the lac operon | ||
| - | ;Lac A | + | ;''Lac A'' |
: Encode for lactose acetylase which transport lactose from medium to ''E.Coli'' | : Encode for lactose acetylase which transport lactose from medium to ''E.Coli'' | ||
| - | ;Lac Y | + | ;''Lac Y'' |
: Encode for lactose permease which regulates concentration lactose inside the cell | : Encode for lactose permease which regulates concentration lactose inside the cell | ||
| - | ;pL | + | ;''pL'' |
: a promoter that can be inhibited by cI | : a promoter that can be inhibited by cI | ||
| Line 132: | Line 131: | ||
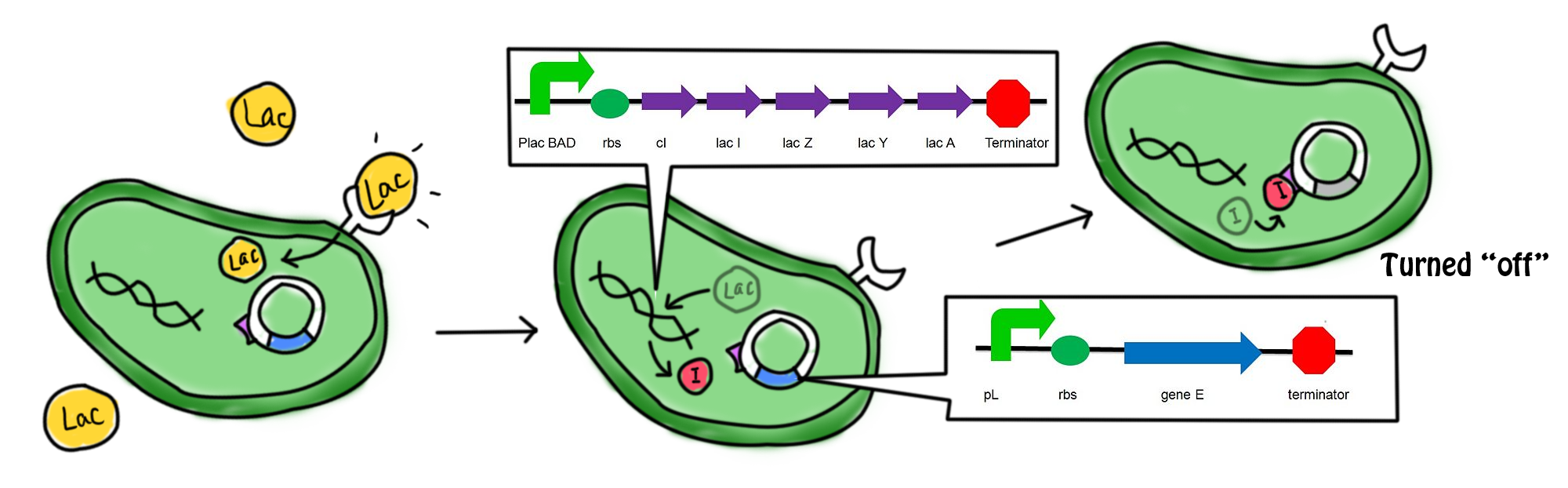
[[Image:Mechaa1.png|700px]] | [[Image:Mechaa1.png|700px]] | ||
| - | *In the presence of lactose, the lac operon and ''cI'' which were originally suppressed by a repressor are now turned on. Transcription, translation and post-modifications are carried out in order to express the gene. cI protein and the β-galactosidase ''(hydrolytic enzyme catalyzing the hydrolysis of lactose)'' are produced. The lactose starts to get digested. The cI protein represses the promoter of suicide gene ''E'', pL, which in turn suppresses the expression of the suicide gene. | + | *In the presence of lactose, the lac operon and ''cI'' which were originally suppressed by a repressor are now turned on. Transcription, translation and post-modifications are carried out in order to express the gene. cI protein and the β-galactosidase ''(hydrolytic enzyme catalyzing the hydrolysis of lactose)'' are produced. The lactose starts to get digested. The cI protein represses the promoter of suicide gene ''E'', ''pL'', which in turn suppresses the expression of the suicide gene. |
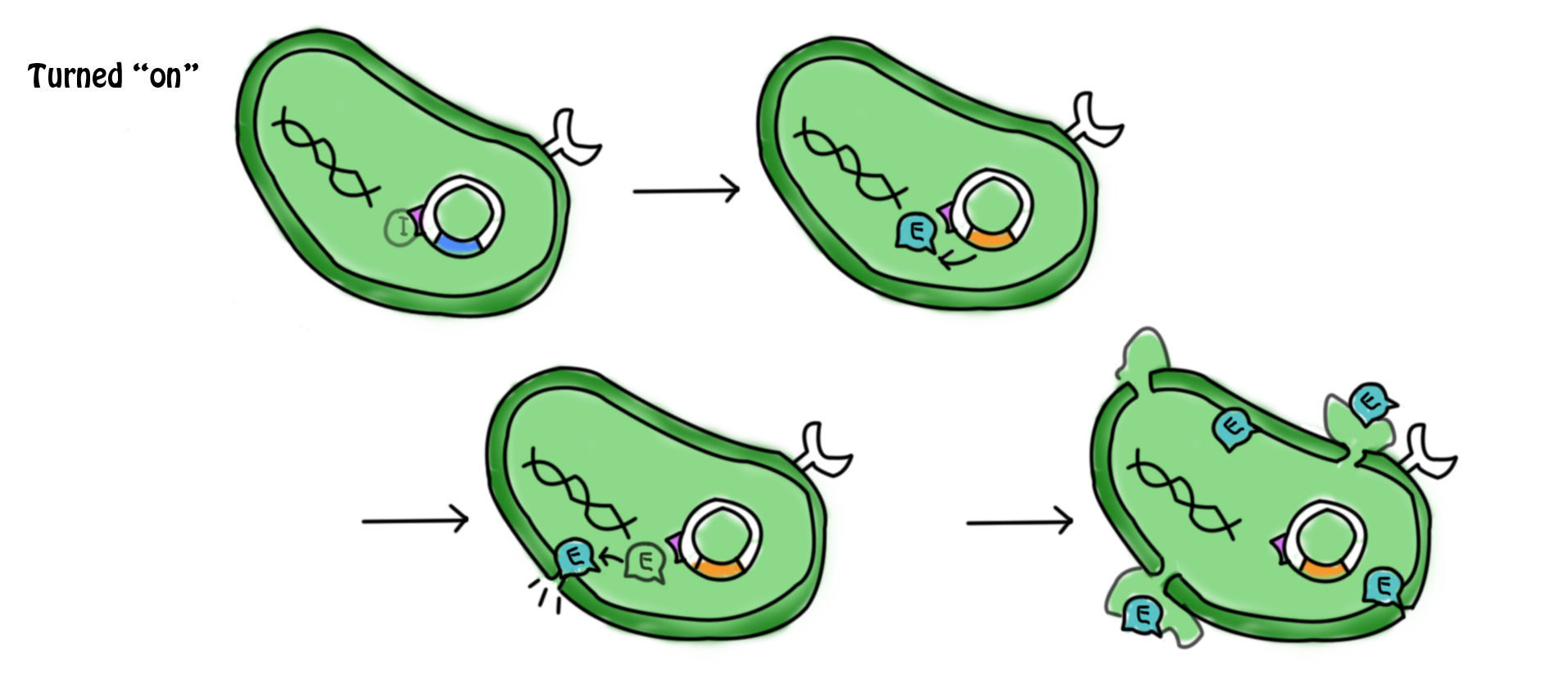
[[Image:Mechaa13.png|700px]] | [[Image:Mechaa13.png|700px]] | ||
| Line 146: | Line 145: | ||
== Results == | == Results == | ||
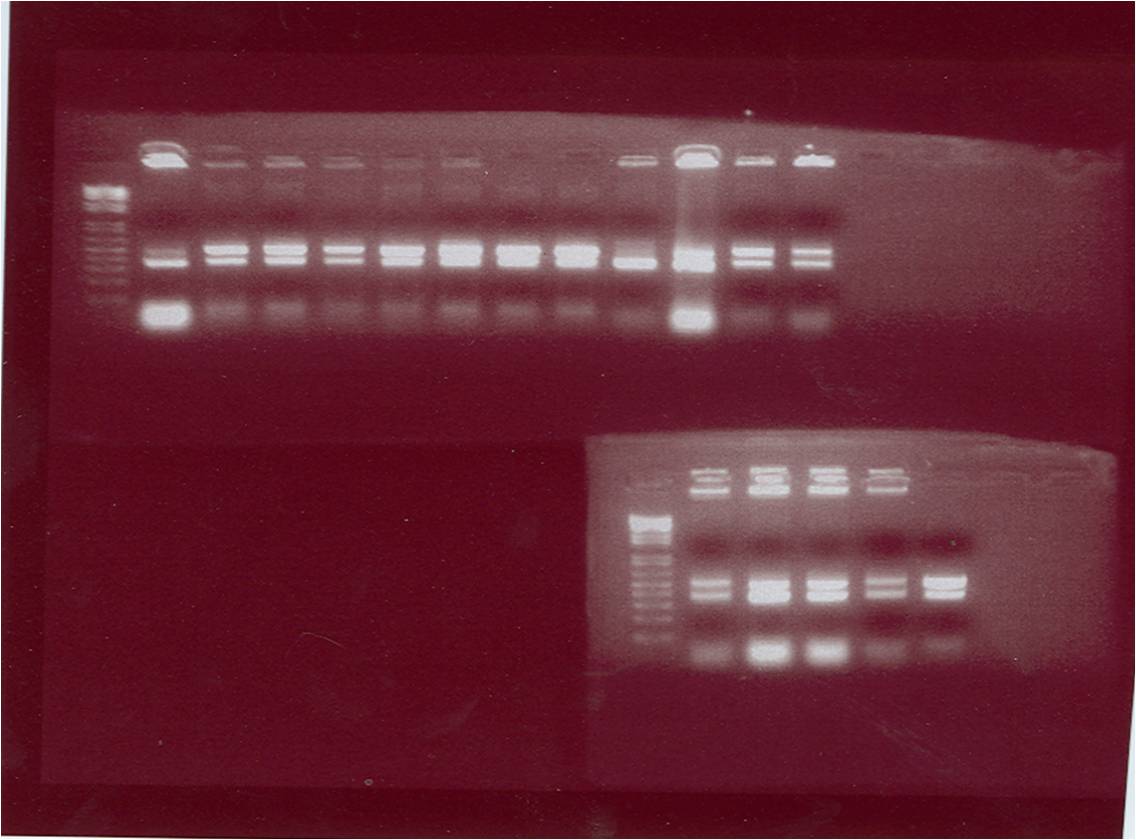
| - | [[Image:gel photo of gene | + | [[Image:gel photo of gene E.png|400px]] [[Image:gel photo of pET28a.png|300px]] |
The figure above shows the gel photo of the suicide gene ''E'' | The figure above shows the gel photo of the suicide gene ''E'' | ||
| - | The gel photo of the plasmid pET28a | + | The gel photo of the plasmid ''pET28a'' |
| - | + | ||
[[Image:OD data.png|430px]] | [[Image:OD data.png|430px]] | ||
Latest revision as of 12:57, 27 October 2010

Contents |
Introduction
After the British Petroleum oil spill that occurred off the coast of the Gulf of Mexico earlier this year, it seemed only natural that our team members take an environmental outlook when it came to brainstorming possible research areas. While researching bacteria capable of oil digestion we found that some varieties are in actual fact pathogenic and so if actually used to clean up oil in the ocean, they may result in harm to the environment or wildlife after completion of their function (degrading oil). Armed with this knowledge, we naturally started to envision a project where we would not just create a solution to the problem of oil spills but also engineer a mechanism that could have potential use in any application of genetic engineering. Apart from bacteria in the degradation of oil we believe that our “bio-safety net” must be implemented into bacteria used in many other applications such as for the killing of cancer cells.
Overall project
Ways to achieve this
- Insertion of killing genes
- Regulated by specific promoters
- Promoters respond to changes in the environment
Hence bacteria can and can only function well and survive in a specific environment
Project Details
Killing Mechanism
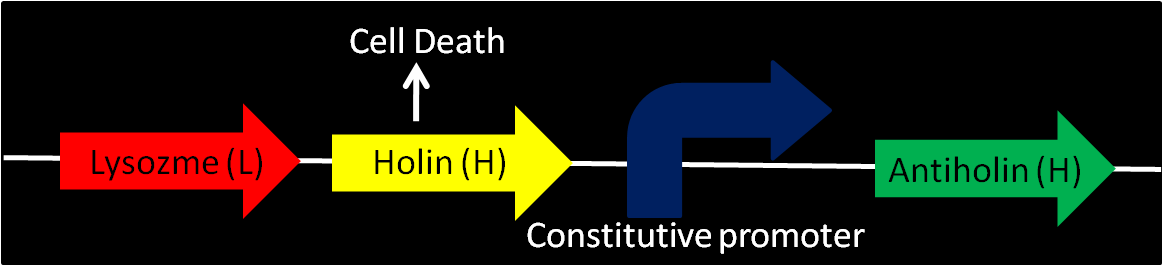
Enterobacteriophage T4 Lysis Device
- We found that The Berkeley's 2008 iGEM team has created a biobrick [Part:BBa_K112808, with no promotor] that could possibly be used as a means to attain cell-lysis.
- Using genes from enterobacteriophage T4 (endolysin-lysozyme, holin and antiholin).
- T4 Holin
- inner membrane-bound proteins, creating pores in the membrane and thus permeabilizing the membrane.
- T4 Antiholin
- binding to and inactivating holins, producing inactive holin-antiholin dimers.
- Lysozyme
- T4 endolysin
- entering the periplasm and degrading peptidoglycan, resulting in cell lysis.
The above picture illustrates the mechanism of the cell-lysis
- As the device comes with no promotor, a variety of promoters could be installed to control cell-lysis.
- It should be pointed out that antiholin has its own constitutive promoter to prevent formation of holin multimers from basal expression. It is of importance as when the device is turned off, relatively higher expression of antiholin is needed and necessary for better stability of the device.
- below is another gene we found it possible to achieve the cell-lysis
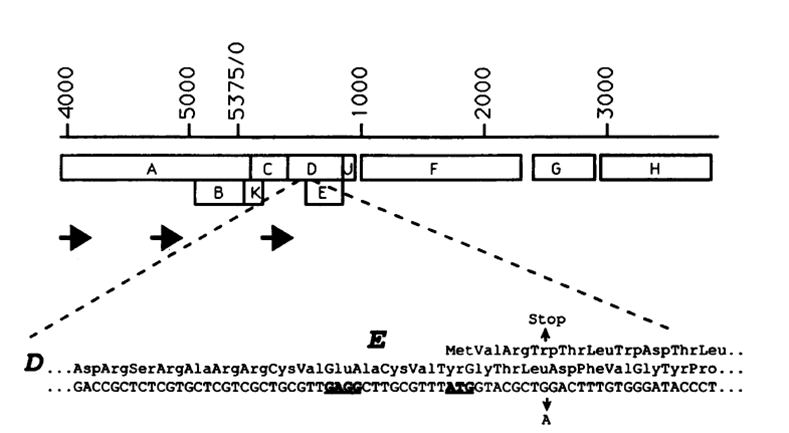
Gene E from Bacteriophage phiX174
The figure shows the location of gene E in the phiX174 genome (YOUNG, MICROBIOLOGIcAL REVIEWS, Sept. 1992,Vol. 56, No. 3)
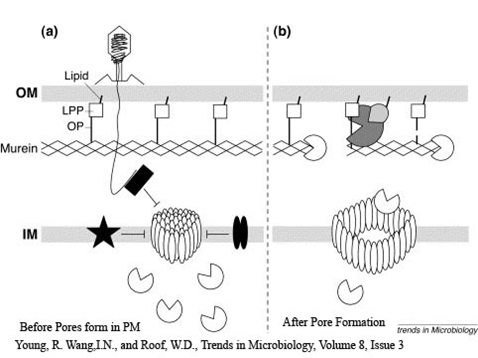
- Gene E is the killing gene in the bacteriophage phiX174. Expression of gene E lead to lysis of the cell.
- Lysis works without elaborating a lytic enzyme, lysozyme
- Small, phase-dark spherical bulges or blebs were developed in host cells
- These blebs are able to grow as large as the summed volumes of the residual rod-shaped portions of the cell.
- Spherical bodies become freed and emptied to become broken ghosts
- They become less refractile and leaving visible membranous debris
- Some blebs burst and other phage are released
- Small lesions which are also called transmembrane tunnels start to appear
- Lesions cause destruction of inner and outer membrane
- Small holes at specific and distinct location in the cell formed
- Sudden collapse of membrane integrity can be caused
- The membrane potential and osmotic gradient across envelop collapsed
Changes in our plans
As a team we decided to make changes to our original goal of creating a bio-safety net to be implemented into an oil degrading bacteria, due to difficulties we encountered and the time constraints on our project.
Difficulties
Oil is a mixture of chemicals composed mainly of hydrocarbons such as alkanes, carboxylic acids and various other aromatic compounds. As a direct result of this crude oil as a whole component cannot be relied upon to induce any promoter, since the promoters available to us may only be induced by certain chemical compounds (not a mixture) and because it is very difficult to target a specific component in crude oil. After assessing the feasibility of using a true oil degrading mechanism, we have concluded that it would be far too difficult to implement such a system into an E. Coli within the time period available to us.
The changes
After identification of the difficulties, we decided to change our plan and instead of focusing on oil, we will use lactose to imitate ‘oil’. This lactose analogue can also be substituted with different subtracts with appropriate modification on the biobricks in initiating the mechanism. Lactose is used as the analogue owing to various reasons:
- Lactose operon is widely studied
- No reporter needed
- Lac-Z assay
- Optimal difficulty and achievable for 2 months’ time
So, now our plan has changed from making an oil digesting bacterium to a lactose digesting bacterium. After the digestion is done, the bacteria will be programmed to kill themselves.
The plasmid design
- PlacBAD
- It is the promoter of the lac operon inducible by lactose or arabinose. Without the substrate, the promoter is suppressed by the repressor protein. Under this condition, the gene is considered as turned off. No expression of genes that are upstream the suppressed promoter can be observed. No proteins that are encoded by these genes are produced.
- cI
- It is the coding sequence of cI protein. The cI protein acts as a repressor protein and bind to the promoter of the suicide gene. The suicide mechanism is then turned off without expression of gene E and production of E protein.
- Lac I
- Encode for the inhibitor of the lac operon
- Lac Z
- Encode for Beta-galactosidase which breakdowns lactose to glucose & galactose and convert lactose to L-lactose which can activate the lac operon
- Lac A
- Encode for lactose acetylase which transport lactose from medium to E.Coli
- Lac Y
- Encode for lactose permease which regulates concentration lactose inside the cell
- pL
- a promoter that can be inhibited by cI
Reasons for choosing the lac operon (since Lactose is now an analog for “crude oil molecules”)
- Found naturally in E.coli
- Well-studied
- Easy to understand how the mechanisms of the operon work
- Present in the chromosomes of E.coli, so that recombination of a new promoter can be easily performed
Scenarios
- In the presence of lactose, the lac operon and cI which were originally suppressed by a repressor are now turned on. Transcription, translation and post-modifications are carried out in order to express the gene. cI protein and the β-galactosidase (hydrolytic enzyme catalyzing the hydrolysis of lactose) are produced. The lactose starts to get digested. The cI protein represses the promoter of suicide gene E, pL, which in turn suppresses the expression of the suicide gene.
- The lactose in the cell culture is continuously digested. After all the lactose has been digested, the promoters for the cI gene and the lac operon are then suppressed. No β-galactozidase and the cI protein are produced. In the absence of cI protein, the suicide gene is turned on. The expression of gene E (production of E protein) leads to the formation of holes in the cell membrane which causes cell death. The mechanism for which can be referred to in the previous section.
- During the culture of the bacteria, arabinose is added to the LB agar (the growth medium for the bacteria). The promoter for the cI and lac operon can also be activated by arabinose. Therefore, cI protein is produced continuously to suppress the expression of the suicide gene. This can prevent bacterial cell death under undesirable conditions.
Possible flaws
- Leaky expression: It might lead to the production of cl proteins which inhibit the expression of the suicide gene. This means that bacteria might still survive after they have finished their function (of digesting all the lactose).
- The suicide genes do not work: Even when there is no inhibition to the suicide gene, the killing mechanism might still not work due to a variety of reasons such as incorrect modifications to proteins, improper folding of the proteins or undesired mutations in the gene.
- The efficacy of the killing mechanism (i.e. time needed for cell-lysis) If the efficacy is too low the bacteria will not die as quickly as we want them to.
Results
The figure above shows the gel photo of the suicide gene E The gel photo of the plasmid pET28a
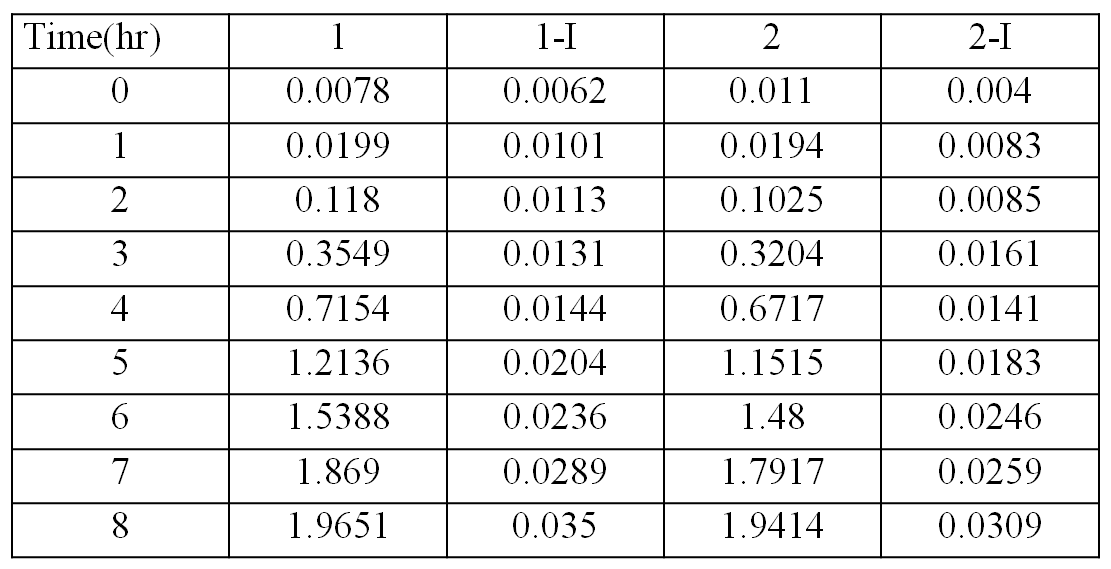
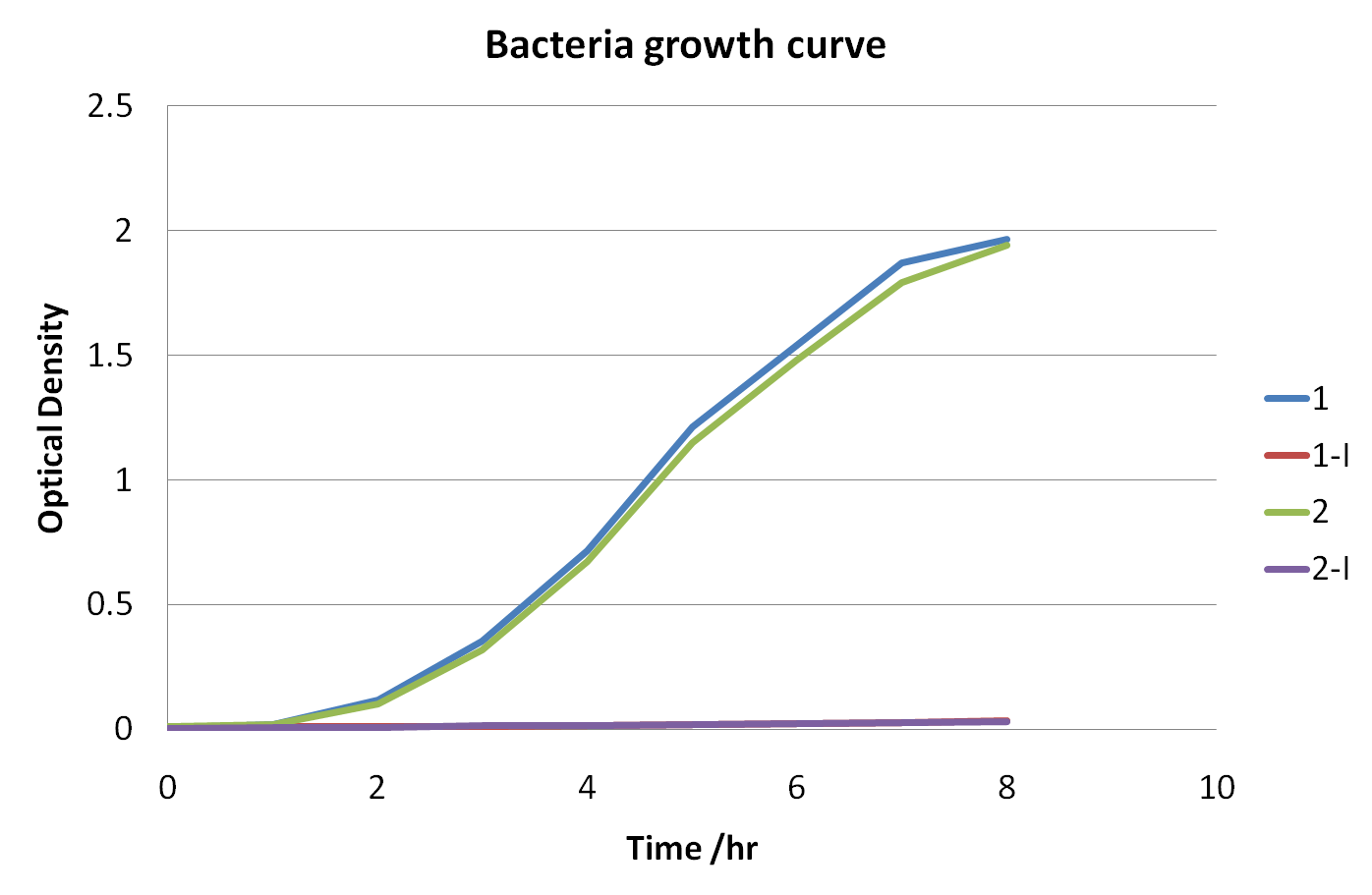
The table shows the optical densities of the bacterial culture with the bacteria contain transformed plasmids
The graph shows the changes in optical density versus time
Optical density measures the concentration of bacteria in suspension, by measuring the amount of light that they collectively absorb. The blue and green curves represent samples taken from bacteria growth of cells where activity of the killing genes was suppressed (i.e. with the presence of arabinose), because of this inhibition the unexpressed genes had no control on cell division and growth resulting in the relatively rapid increases to the curve. The curves that appear more flat are those where the expression of the killing genes were induced (by the presence of IPTG) meaning that cell lysis occurred, resulting in cell death. These curves show very low optical densities which indicate almost nonexistent bacterial growth in those samples, meaning the killing genes successfully work.
Possible further research
- A key area for future research would be to engineer the plasmid in such a way that promoters could be more readily interchangeable, so that the killing genes can be induced by molecules relevant to their purposes (e.g. heavy metals or rganic materials).
- With interchangeable promoters we would also need to make chosen bacteria code for certain proteins that are specific to intended functions (e.g. cancer digesting proteins from bacteria).
- By making it easier for the promoters and coded proteins to be selected individually, the “bio-safety net” would become a universal safety mechanism that could be incorporated into engineered bacteria performing any function, and would be regulated by a user-defined substance.
 "
"