Team:NYMU-Taipei/Project/Speedy switch
From 2010.igem.org
Barry50710 (Talk | contribs) (→Abstract) |
Blackrabbit (Talk | contribs) (→Experiments) |
||
| (123 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
| - | == | + | =<font color=blue>Abstract</font>= |
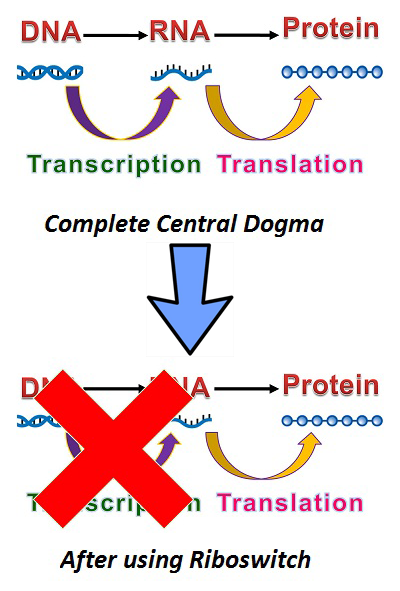
| - | [[Image:NYMU_Central_Dogma.png|thumb|right|The Central Dogma (top) and the effective central dogma when using riboswitches (bottom). Transcription is effectively skipped when ready for use, since the mRNA will have already been expressed.]] | + | [[Image:NYMU_Central_Dogma.png|thumb|250px|right|The Central Dogma (top) and the effective central dogma when using riboswitches (bottom). Transcription is effectively skipped when ready for use, since the mRNA will have already been expressed.]] |
| - | + | '''Speedy Switch controls the "on/off " of RNA translation. We use "Riboswitch" as our speedy switch to pause central dogma at the mRNA level.''' | |
| - | + | ||
| - | == | + | |
| + | In the past, translating proteins from DNA has followed the central dogma of molecular biology: DNA to RNA to Protein. Normally, after mRNA is transcribed from DNA, ribosomes will bind to the ribosome binding site(RBS) and begin translating mRNA into protein. During this process, we have no way of knowing the location, nor the quantity of mRNA; and after the process, mRNA is quickly degraded. As such, it is very hard to research the detailed roles and implications of mRNA in the central dogma. To resolve this problem, we placed a mRNA level based switch which can be used to control the translation of mRNA: riboswitch. | ||
| + | |||
| + | =<font color=blue>Introduction</font>= | ||
[[Image:NYMU Pre-ribo2.jpg|thumb|right|250px|The riboswitch is turned off. The RBS is hidden by the secondary structure the riboswitch forms. The ribosome is unable to bind to the RBS, suspending translation of the downstream gene.]] | [[Image:NYMU Pre-ribo2.jpg|thumb|right|250px|The riboswitch is turned off. The RBS is hidden by the secondary structure the riboswitch forms. The ribosome is unable to bind to the RBS, suspending translation of the downstream gene.]] | ||
[[Image:NYMU Ribo-2.jpg|thumb|right|250px|The riboswitch is turned on when a specific small molecule binds to it. The ribosome can then bind to the RBS, inducing translation of the downstream gene.]] | [[Image:NYMU Ribo-2.jpg|thumb|right|250px|The riboswitch is turned on when a specific small molecule binds to it. The ribosome can then bind to the RBS, inducing translation of the downstream gene.]] | ||
| - | |||
| - | + | '''Function of Speedy Switch: speed up the cycle from DNA to mRNA to protein and act as a switch between the mRNA and protein levels''' | |
| - | + | In our project, Speedy Switch serves two main roles. The first is to speed up the expression cycle from DNA to mRNA to protein. Using a riboswitch we can pretranscribe a DNA into an mRNA, ready to be translated at a moments notice. In essence, we can produce protein without having to wait for transcription. The second role the riboswitch serves is to act as a switch between the mRNA and protein levels. Using a riboswitch, we can regulate downstream mRNA and control the expression of proteins. | |
| - | + | ||
| - | + | ||
| - | + | Typically, since translation often occurs at the moment mRNA passed into the cytoplasm, protein and mRNA normally exist together. With a riboswitch control, we can study both the expression of mRNA and the expression of protein in the same cells, without protein-mRNA interference. | |
| - | |||
| - | + | '''We use "Riboswitch" as our speedy switch, which can control translation. It can be divided into two parts: a sensor and an actuator.''' | |
| - | + | Before the discovery of RNA regulatory system , the only way to induce reaction in a cell was through inducible promoters. By turning these promoters on or off, we could control the transcription of the downstream DNA into RNA thus also controlling the translation of RNA to Protein. However, with only promoters we traditionally skip the RNA system involved in the pathway of protein synthesis. By inserting a "switch" between the DNA and RNA system we can make a thorough inspection into the individual mechanism of both systems and the cross-effect between their regulatory factors. | |
| - | *Verification of protein function | + | |
| + | The discovery of the riboswitch was based on data which described conserved mRNA secondary structure found on 5’-untranslated regions and the creation of small-molecule binding mRNA, sensors. The function of these riboswitches is similar to the function of inducible promoters in that they both regulate downstream genetic data: their difference is that while promoters regulate transcription of DNA, riboswitches control translation of mRNA. | ||
| + | |||
| + | A riboswitch is a part of mRNA molecule that can bind a small molecule. When it does, the riboswitch will change its structure to regulate the following gene's activity. | ||
| + | |||
| + | A riboswitch has two parts: a sensor and an actuator. These two components work together to form a ‘switch’. The sensor binds to a small molecule inducer, and the actuator structurally changes to regulate gene expression. | ||
| + | (Harbaugh et al.,2008)(Lynch et al., 2006) | ||
| + | ==Purpose== | ||
| + | *Verification of protein function | ||
| + | **We can perform RNA assay and protein assay in the same cell | ||
*Control of protein expression | *Control of protein expression | ||
| - | =====Design | + | =<font color=blue>Experiments</font>= |
| - | In order for a riboswitch to work in our | + | |
| + | [[Team:NYMU-Taipei/Experiments/Speedy_switch|Reporter Gene Assay Experiment Data]] show that the Theophylline Riboswitch works and has an increased rate of transcription (up to a point) with an increased amount of Theophylline. | ||
| + | |||
| + | ==Design== | ||
| + | In order for a suitable riboswitch to work in our experiments, it needs to have the following characteristics: | ||
* The inducer does not naturally exist or metabolize in the target organism. | * The inducer does not naturally exist or metabolize in the target organism. | ||
* The riboswitch does not exist naturally in the target organism. | * The riboswitch does not exist naturally in the target organism. | ||
| Line 34: | Line 45: | ||
** Although we can modify the cutting sites of our riboswitch, this action may cause more problems: the cutting site may mutate the secondary structure and molecule binding sites causing it to cease function. | ** Although we can modify the cutting sites of our riboswitch, this action may cause more problems: the cutting site may mutate the secondary structure and molecule binding sites causing it to cease function. | ||
| - | + | ===An example of our '''Speedy Switch'''=== | |
| - | + | Because we were using ''Escherichia coli'' DH5alpha as our chassie organism, so we decided to use '''Theophylline Riboswitch''' which fits all these requirements for our circuit design and enginnering. | |
| - | + | ||
| - | + | ||
| - | + | Since most riboswitches already have Ribosome binding sites (RBSs) in their structures, we did not add other RBSs in front of downstream reporters. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | After transforming the whole structure,"promoter+ riboswitch+ GFP+ terminator in plasmid" into ''Escherichia coli,'' | |
| - | + | the cells will express GFP when theophylline (the inducer) is introduced. | |
| - | [[Image:NYMU Ribo circuit.jpg | + | ==Composition of our circuit== |
| + | [[Image:NYMU Ribo circuit.jpg|left|thumb|500px|This is our circuit of speedy switch. Theophylline Riboswitch sequence is after the promoter, inclusive of the ribosome binding site. And then we put GFP,next is the terminator. We just add Theophylline, GFP will emit green fluorescence.]] | ||
| Line 69: | Line 76: | ||
| - | |||
| - | |||
To test our hyposthesis, we needed to construct a circuit that has a promoter, a riboswitch, a reporter, and a terminator. We chose to use the theophylline riboswitch as it suited our requirements. | To test our hyposthesis, we needed to construct a circuit that has a promoter, a riboswitch, a reporter, and a terminator. We chose to use the theophylline riboswitch as it suited our requirements. | ||
| Line 76: | Line 81: | ||
When the full sequence outlined above is transformed into the bacteria, it waits, inactivated, for the right small molecule inducer, in this case, theophylline. When theophylline is added, it will induce the riboswitch to fold differently to allow the translation of the downstream gene, without waiting for transcription. | When the full sequence outlined above is transformed into the bacteria, it waits, inactivated, for the right small molecule inducer, in this case, theophylline. When theophylline is added, it will induce the riboswitch to fold differently to allow the translation of the downstream gene, without waiting for transcription. | ||
| - | By comparing the flourescence intensity data (the speed of GFP production), we can determine the difference in time between the traditional method of inducing promoters, to our method of inducing mRNA. | + | By comparing the flourescence intensity data (the speed of GFP production), we can determine the difference in time between the traditional method of inducing promoters, to our method of inducing mRNA. |
| + | ===Promoter=== | ||
| + | *We used '''pLac''' constitutive promoter from Biobrick Parts Registry{{:Team:NYMU-Taipei/BBa|R0010}} | ||
| + | We used this constitutive promoter because it can keep working without additional stimulation. So we can get sufficient mRNA with Theophylline Riboswitch in ''Escherichia coli'' DH5alpha. We just need to add Theophylline and Theophylline Riboswitch will function rapidly. | ||
<!-- * Although we have the sequence for the theophylline riboswitch, without a template, we could not run PCR (Polymerase Chain Reaction) to produce the quantity of DNA we required. To create this template, we design forward and reverse primers that have a common region. | <!-- * Although we have the sequence for the theophylline riboswitch, without a template, we could not run PCR (Polymerase Chain Reaction) to produce the quantity of DNA we required. To create this template, we design forward and reverse primers that have a common region. | ||
--> | --> | ||
| - | Since the sequence length of this riboswitch is relatively short, we decided to synthesize the riboswitch directly using two primers (which also contain the biobrick prefix and suffix) | + | |
| + | ===Theophylline Riboswitch=== | ||
| + | Since the sequence length of this riboswitch is relatively short, we decided to synthesize the riboswitch directly using two primers (which also contain the biobrick prefix and suffix) | ||
*sequence | *sequence | ||
ggtgataccagcatcgtcttgatgcccttggcagcaccccgctgcaagacaacaag | ggtgataccagcatcgtcttgatgcccttggcagcaccccgctgcaagacaacaag | ||
| Line 92: | Line 102: | ||
gtcttgatgcccttggcagcaccccgctgcaagacaacaagtactagtagcggccgctgcag | gtcttgatgcccttggcagcaccccgctgcaagacaacaagtactagtagcggccgctgcag | ||
| - | These two primers anneal | + | These two primers anneal at this common region. |
| + | |||
| + | ===GFP+terminator=== | ||
| + | *We used biobrick {{:Team:NYMU-Taipei/BBa|J04630}} | ||
| + | We used Green fluorescent protein as our reporter for two main reasons. First, GFP makes a great reporter because it fluoresces when it is activated, making itself easy to detect. We can measure the activity of the promoter by the intensity of the fluorescence . The second reason is that the GFP used is a biobrick, thus if another team needs to use this riboswitch circuit, it would be easy for them to attach another biobrick. So we chose biobrick - {{:Team:NYMU-Taipei/BBa|J04630}} (GFP with terminator) | ||
| + | |||
| + | ==Whole Process== | ||
| + | # First, two riboswitch primers will anneal together at the common region through PCR amplification. | ||
# We then digested the riboswitch PCR product and a plasmid containing the plasmid backbone pSB1A2 with the restriction enzymes XbaI and PstI. | # We then digested the riboswitch PCR product and a plasmid containing the plasmid backbone pSB1A2 with the restriction enzymes XbaI and PstI. | ||
# After gel extraction/PCR purification of the relevant parts, we ligated them and produced the biobrick part {{:Team:NYMU-Taipei/BBa|K411101}}. | # After gel extraction/PCR purification of the relevant parts, we ligated them and produced the biobrick part {{:Team:NYMU-Taipei/BBa|K411101}}. | ||
# Performed a back insert of {{:Team:NYMU-Taipei/BBa|J04630|(GFP+terminator)}} (digested with XbaI and PstI) into {{:Team:NYMU-Taipei/BBa|K411101}} (digested with SpeI and PstI) and formed the biobrick {{:Team:NYMU-Taipei/BBa|K411102}}. | # Performed a back insert of {{:Team:NYMU-Taipei/BBa|J04630|(GFP+terminator)}} (digested with XbaI and PstI) into {{:Team:NYMU-Taipei/BBa|K411101}} (digested with SpeI and PstI) and formed the biobrick {{:Team:NYMU-Taipei/BBa|K411102}}. | ||
# Performed another back insert of {{:Team:NYMU-Taipei/BBa|K411102}} (digested with XbaI and PstI) into {{:Team:NYMU-Taipei/BBa|R0010|(lac promoter)}} (digested with SpeI and PstI) and formed the biobrick {{:Team:NYMU-Taipei/BBa|K411103}}. | # Performed another back insert of {{:Team:NYMU-Taipei/BBa|K411102}} (digested with XbaI and PstI) into {{:Team:NYMU-Taipei/BBa|R0010|(lac promoter)}} (digested with SpeI and PstI) and formed the biobrick {{:Team:NYMU-Taipei/BBa|K411103}}. | ||
| - | + | # Finally we tested this kind of ''Escherichia coli''. We add Theophylline to induce riboswitch and and translate GFP. | |
| + | |||
| + | =<font color=blue>Result</font>= | ||
| + | In theory, if we add more inducers to a cell, riboswitch will translate more GFPs. But theophylline is toxic for ''Escherichia coli.'' From paper, we know that if theophylline concentration is over 5 μM, ''Escherichia coli'' will die.(Lynch et al., 2006) | ||
| + | [[Image:20101025(org).png|800px|In this picture, it shows that when more Theophylline is added, more GFP is created and fluoresces. | ||
| + | For more of our speedy switch experimental data, please check out our SpeedBac experimental result section.]] | ||
| + | |||
| + | This figure shows the results of our experiments. Comparing to N (stands for the Negative control), we find GFP express when we add Theophylline. | ||
| + | For more Speedy Switch experimental data, please check out our SpeedBac experimental results [[Team:NYMU-Taipei/Experiments/Speedy switch | here]]. | ||
| + | |||
| + | =<font color=blue>Reference</font>= | ||
| + | *Desai, S.K., and Gallivan, J.P. (2004). Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. ''J Am Chem Soc'' 126, 13247-13254. | ||
| + | |||
| + | *Harbaugh S.V., Davidson M.E., Chushak Y.G., Kelley-Loughnane N., and Stone M.O.(2008) Riboswitch-based sensor in low optical background. ''Proc. of SPIE'' 7040, 70400C | ||
| + | |||
| + | *Lynch, S.A., Desai, S.K., Sajja, H.K., and Gallivan, J.P. (2007). A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. ''Chem Biol'' 14, 173-184. | ||
| + | |||
| + | *Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., and Breaker, R.R. (2003). Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. ''Cell'' 113, 577-586. | ||
| + | |||
| + | *Suess, B., Fink, B., Berens, C., Stentz, R., and Hillen, W. (2004). A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. ''Nucleic Acids Res'' 32, 1610-1614. | ||
| - | + | *Topp, S., and Gallivan, J.P. (2007). Guiding bacteria with small molecules and RNA. ''J Am Chem Soc'' 129, 6807-6811. | |
| - | + | ||
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Latest revision as of 23:51, 27 October 2010
| Home | Project Overview | Speedy reporter | Speedy switch | Speedy protein degrader | Experiments and Parts | Applications | F.A.Q | About Us |
Contents |
Abstract
Speedy Switch controls the "on/off " of RNA translation. We use "Riboswitch" as our speedy switch to pause central dogma at the mRNA level.
In the past, translating proteins from DNA has followed the central dogma of molecular biology: DNA to RNA to Protein. Normally, after mRNA is transcribed from DNA, ribosomes will bind to the ribosome binding site(RBS) and begin translating mRNA into protein. During this process, we have no way of knowing the location, nor the quantity of mRNA; and after the process, mRNA is quickly degraded. As such, it is very hard to research the detailed roles and implications of mRNA in the central dogma. To resolve this problem, we placed a mRNA level based switch which can be used to control the translation of mRNA: riboswitch.
Introduction
Function of Speedy Switch: speed up the cycle from DNA to mRNA to protein and act as a switch between the mRNA and protein levels
In our project, Speedy Switch serves two main roles. The first is to speed up the expression cycle from DNA to mRNA to protein. Using a riboswitch we can pretranscribe a DNA into an mRNA, ready to be translated at a moments notice. In essence, we can produce protein without having to wait for transcription. The second role the riboswitch serves is to act as a switch between the mRNA and protein levels. Using a riboswitch, we can regulate downstream mRNA and control the expression of proteins.
Typically, since translation often occurs at the moment mRNA passed into the cytoplasm, protein and mRNA normally exist together. With a riboswitch control, we can study both the expression of mRNA and the expression of protein in the same cells, without protein-mRNA interference.
We use "Riboswitch" as our speedy switch, which can control translation. It can be divided into two parts: a sensor and an actuator.
Before the discovery of RNA regulatory system , the only way to induce reaction in a cell was through inducible promoters. By turning these promoters on or off, we could control the transcription of the downstream DNA into RNA thus also controlling the translation of RNA to Protein. However, with only promoters we traditionally skip the RNA system involved in the pathway of protein synthesis. By inserting a "switch" between the DNA and RNA system we can make a thorough inspection into the individual mechanism of both systems and the cross-effect between their regulatory factors.
The discovery of the riboswitch was based on data which described conserved mRNA secondary structure found on 5’-untranslated regions and the creation of small-molecule binding mRNA, sensors. The function of these riboswitches is similar to the function of inducible promoters in that they both regulate downstream genetic data: their difference is that while promoters regulate transcription of DNA, riboswitches control translation of mRNA.
A riboswitch is a part of mRNA molecule that can bind a small molecule. When it does, the riboswitch will change its structure to regulate the following gene's activity.
A riboswitch has two parts: a sensor and an actuator. These two components work together to form a ‘switch’. The sensor binds to a small molecule inducer, and the actuator structurally changes to regulate gene expression. (Harbaugh et al.,2008)(Lynch et al., 2006)
Purpose
- Verification of protein function
- We can perform RNA assay and protein assay in the same cell
- Control of protein expression
Experiments
Reporter Gene Assay Experiment Data show that the Theophylline Riboswitch works and has an increased rate of transcription (up to a point) with an increased amount of Theophylline.
Design
In order for a suitable riboswitch to work in our experiments, it needs to have the following characteristics:
- The inducer does not naturally exist or metabolize in the target organism.
- The riboswitch does not exist naturally in the target organism.
- The riboswitch does not have EcoRI, XbaI, SpeI, or PstI cutting sites.
- Although we can modify the cutting sites of our riboswitch, this action may cause more problems: the cutting site may mutate the secondary structure and molecule binding sites causing it to cease function.
An example of our Speedy Switch
Because we were using Escherichia coli DH5alpha as our chassie organism, so we decided to use Theophylline Riboswitch which fits all these requirements for our circuit design and enginnering.
Since most riboswitches already have Ribosome binding sites (RBSs) in their structures, we did not add other RBSs in front of downstream reporters.
After transforming the whole structure,"promoter+ riboswitch+ GFP+ terminator in plasmid" into Escherichia coli, the cells will express GFP when theophylline (the inducer) is introduced.
Composition of our circuit
To test our hyposthesis, we needed to construct a circuit that has a promoter, a riboswitch, a reporter, and a terminator. We chose to use the theophylline riboswitch as it suited our requirements.
When the full sequence outlined above is transformed into the bacteria, it waits, inactivated, for the right small molecule inducer, in this case, theophylline. When theophylline is added, it will induce the riboswitch to fold differently to allow the translation of the downstream gene, without waiting for transcription.
By comparing the flourescence intensity data (the speed of GFP production), we can determine the difference in time between the traditional method of inducing promoters, to our method of inducing mRNA.
Promoter
- We used pLac constitutive promoter from Biobrick Parts Registry[http://partsregistry.org/Part:BBa_R0010 BBa_R0010]
We used this constitutive promoter because it can keep working without additional stimulation. So we can get sufficient mRNA with Theophylline Riboswitch in Escherichia coli DH5alpha. We just need to add Theophylline and Theophylline Riboswitch will function rapidly.
Theophylline Riboswitch
Since the sequence length of this riboswitch is relatively short, we decided to synthesize the riboswitch directly using two primers (which also contain the biobrick prefix and suffix)
- sequence
ggtgataccagcatcgtcttgatgcccttggcagcaccccgctgcaagacaacaag forward primer : gaattcgcggccgcttctagag ggtgataccagcatcgtcttgatgcccttggcag reverse primer : ctgcagcggccgctactagtacttgttgtcttgcagcggggtgctgccaagggcatcaagac
PCR expected result (99bp)
gaattcgcggccgcttctagagggtgataccagcatcgtcttgatgcccttggcag
gaattcgcggccgcttctagagggtgataccagcatcgtcttgatgcccttggcagcaccccgctgcaagacaacaagtactagtagcggccgctgcag
gtcttgatgcccttggcagcaccccgctgcaagacaacaagtactagtagcggccgctgcag
These two primers anneal at this common region.
GFP+terminator
- We used biobrick [http://partsregistry.org/Part:BBa_J04630 BBa_J04630]
We used Green fluorescent protein as our reporter for two main reasons. First, GFP makes a great reporter because it fluoresces when it is activated, making itself easy to detect. We can measure the activity of the promoter by the intensity of the fluorescence . The second reason is that the GFP used is a biobrick, thus if another team needs to use this riboswitch circuit, it would be easy for them to attach another biobrick. So we chose biobrick - [http://partsregistry.org/Part:BBa_J04630 BBa_J04630] (GFP with terminator)
Whole Process
- First, two riboswitch primers will anneal together at the common region through PCR amplification.
- We then digested the riboswitch PCR product and a plasmid containing the plasmid backbone pSB1A2 with the restriction enzymes XbaI and PstI.
- After gel extraction/PCR purification of the relevant parts, we ligated them and produced the biobrick part [http://partsregistry.org/Part:BBa_K411101 BBa_K411101].
- Performed a back insert of [http://partsregistry.org/Part:BBa_J04630 BBa_J04630(GFP+terminator)] (digested with XbaI and PstI) into [http://partsregistry.org/Part:BBa_K411101 BBa_K411101] (digested with SpeI and PstI) and formed the biobrick [http://partsregistry.org/Part:BBa_K411102 BBa_K411102].
- Performed another back insert of [http://partsregistry.org/Part:BBa_K411102 BBa_K411102] (digested with XbaI and PstI) into [http://partsregistry.org/Part:BBa_R0010 BBa_R0010(lac promoter)] (digested with SpeI and PstI) and formed the biobrick [http://partsregistry.org/Part:BBa_K411103 BBa_K411103].
- Finally we tested this kind of Escherichia coli. We add Theophylline to induce riboswitch and and translate GFP.
Result
In theory, if we add more inducers to a cell, riboswitch will translate more GFPs. But theophylline is toxic for Escherichia coli. From paper, we know that if theophylline concentration is over 5 μM, Escherichia coli will die.(Lynch et al., 2006)
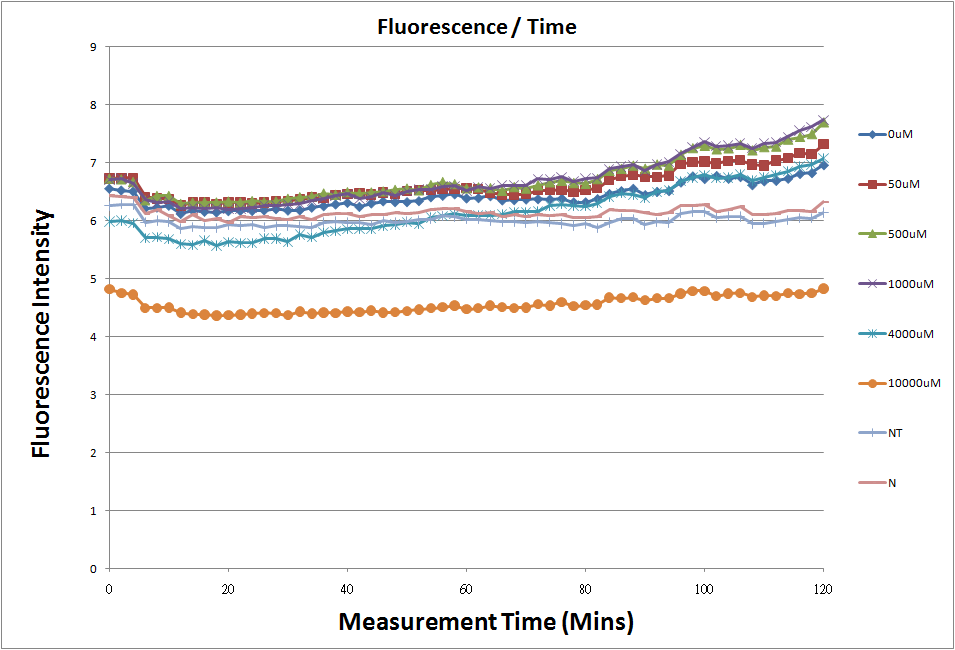
This figure shows the results of our experiments. Comparing to N (stands for the Negative control), we find GFP express when we add Theophylline. For more Speedy Switch experimental data, please check out our SpeedBac experimental results here.
Reference
- Desai, S.K., and Gallivan, J.P. (2004). Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc 126, 13247-13254.
- Harbaugh S.V., Davidson M.E., Chushak Y.G., Kelley-Loughnane N., and Stone M.O.(2008) Riboswitch-based sensor in low optical background. Proc. of SPIE 7040, 70400C
- Lynch, S.A., Desai, S.K., Sajja, H.K., and Gallivan, J.P. (2007). A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem Biol 14, 173-184.
- Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., and Breaker, R.R. (2003). Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577-586.
- Suess, B., Fink, B., Berens, C., Stentz, R., and Hillen, W. (2004). A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res 32, 1610-1614.
- Topp, S., and Gallivan, J.P. (2007). Guiding bacteria with small molecules and RNA. J Am Chem Soc 129, 6807-6811.
 "
"