Team:UT-Tokyo/Sudoku assay MS2
From 2010.igem.org
(New page: __NOTOC__{{UT-Tokyo_CSS3}}{{UT-Tokyo_CSS_p}}{{UT-Tokyo_Head}} = '''Sudoku''' = Introduction [https://2010.igem.org/Team:UT-Tokyo/Sudoku_construct System] [https://2010.igem.org/Team...) |
(→3. Expression test in receiver E.coli) |
||
| (13 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
= '''Sudoku''' = | = '''Sudoku''' = | ||
| - | + | <html> | |
| - | + | <div> | |
| - | + | <ul id="inpagemenu"> | |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_abstract" id="abstract">Introduction</a></li> | |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_construct" id="construct">System</a></li> | |
| - | [https://2010.igem.org/Team:UT-Tokyo/ | + | <li><a href="/Team:UT-Tokyo/Sudoku_modeling" id="modeling">Modeling</a></li> |
| - | [https://2010.igem.org/Team:UT-Tokyo/ | + | <li><span>Experiments</span></li> |
| - | + | <li><a href="/Team:UT-Tokyo/Sudoku_perspective" id="perspective">Perspective</a></li> | |
| + | <li><a href="/Team:UT-Tokyo/Sudoku_reference" id="reference">Reference</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div id="clear"></div> | ||
| + | </html> | ||
| + | <br /> | ||
| + | =>-Assay- | ||
| + | [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_LeakSw Terminator Leak]/ | ||
| + | [https://2010.igem.org/Team:UT-Tokyo/Sudoku_assay_LocSq Location Sequence]/ | ||
| + | Phage MS2 | ||
='''Phage MS2'''= | ='''Phage MS2'''= | ||
| - | ==''' | + | =='''Introduction'''== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | In the signal transmission assay: MS2 phage, we test whether MS2 phage can function as a signal transducer (what we call “Signal Virus”) and determine the conditions under which they function. In other words, we test whether MS2 phage can carry out from bacteria mRNA which codes for number information, infect other ''E.coli'', and whether the mRNA it carries can be translated in the infected E.coli. This signal transmission system requires the possession of 3 properties. In this assay we test for the presence of these properties. First, we test whether functional viruses are produced from MS2 phage cDNA although the MS2 phage genome is never transcribed from DNA in its natural lifecycle. Second, we test whether the targeted mRNA is carried outside ''E.coli''. A wild-type phage genome contains 19bp of loading sequence, around which coat proteins of MS2 phage aggregate. Therefore we test whether the targeted mRNA is carried outside by adding a loading sequence. Third, we test whether viral remnants lacking their natural genome but instead possess the targeted mRNA are able to infect other ''E.coli'' and whether we can detect translation. In this assay, we select the Cre generator and the Gfp generator as the targeted mRNA. If we confirm these three work, we can say our MS2 phage system functions as Signal Virus. | ||
| - | ''' | + | =='''1. Titer test of wild type MS2 phage'''== |
| - | + | [[Image:MS2_Fig1.png|200px|thumb|Figure 1: The construct of experiment 1]] | |
| - | + | In this experiment, we test whether cDNA of MS2 phage genome produces functional viruses. We transform the plasmid coding for the MS2 phage genome into DE3, and induce its expression by T7 RNA polymerase. After that we collect the cell lysate and perform a titer test by infecting JM109 (Figure 1). | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''' | + | =='''2. Measurement of quantity of the targeted mRNA'''== |
| - | + | [[Image:MS2_Fig2.png|200px|thumb|Figure 2: The construct of experiment 2]] | |
| - | + | ||
| - | + | ||
| - | 2 | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | In this experiment we test whether the targeted mRNA is packaged by MS2 coat proteins and carried out of the cell. We use the construct shown in Figure 2. We transform the plasmids coding MS2 phage genome to DE3 cells and induce expression using the T7 polymerase. At the same time we induce transcription from the Gfp generator or The Cre generator each with a loading sequence attached. Then the mRNA is packaged and carried out of the cell. After that we collect the cell lysate and the viral remnants it contains. Next we measure the quantity of mRNA coding for the gfp generator or cre generator. It is possible that the detected mRNA is derived from the cytosol of ''E. coli'' instead of that derived from the virus. In order to quantify cytosolic RNA, we use as control E.coli transformed with cDNA of MS2 phage and the cre generator or gfp generator and quantify the mRNA coding for the cre generator and gfp generator using the same methods. The difference between the samples and the control represents the quantity of mRNA carried out from the cells because mRNA detected under this condition should be derived from the cytosol. We expect the detection of mRNA carried outside by the viral remnants. | ||
| - | |||
| + | =='''3. Expression test in receiver ''E.coli'''''== | ||
| + | [[Image:MS2_Fig3.png|200px|thumb|Figure 3: The construct of experiment 3]] | ||
| + | In this experiment we test whether viral remnants lacking their natural genome but instead possess the targeted mRNA are able to infect other ''E.coli'' and whether we can detect translation. We use the construct shown in Figure 3. We transform the plasmids coding MS2 phage genom e to DE3 cells and induce expression using the T7 polymerase. At the same time we induce transcription from the Gfp generator or The Cre generator each with a loading sequence attached. Then the mRNA is packaged and carried out of the cell. After that we collect the cell lysate and the viral remnants it contains. And we make concentrated phage solution. We test whether we can detect Gfp is translated in the infected ''E.coli''. On the other hand as the receiver, we prepare ''E.coli'' which can express gfp because the terminator is removed by Cre, and infect phage to it. Although gfp can be expressed on both systems, the intensity of gfp emission will not be detected at the same time because in many cases just a few phages can be infected by ''E.coli''. When the gfp expression is detected on either one of the systems, we can conclude that phage in the transmitter ''E.coli'' carry out target mRNA and infect it to other bacteria, and in the result, target mRNA is translated in the infected ''E.coli''. The construct of Hin is prepared for negative-control for gfp generator and cre generator. | ||
| + | |||
{{UT-Tokyo_Foot}} | {{UT-Tokyo_Foot}} | ||
Latest revision as of 03:43, 28 October 2010


Sudoku
=>-Assay- Terminator Leak/ Location Sequence/ Phage MS2
Phage MS2
Introduction
In the signal transmission assay: MS2 phage, we test whether MS2 phage can function as a signal transducer (what we call “Signal Virus”) and determine the conditions under which they function. In other words, we test whether MS2 phage can carry out from bacteria mRNA which codes for number information, infect other E.coli, and whether the mRNA it carries can be translated in the infected E.coli. This signal transmission system requires the possession of 3 properties. In this assay we test for the presence of these properties. First, we test whether functional viruses are produced from MS2 phage cDNA although the MS2 phage genome is never transcribed from DNA in its natural lifecycle. Second, we test whether the targeted mRNA is carried outside E.coli. A wild-type phage genome contains 19bp of loading sequence, around which coat proteins of MS2 phage aggregate. Therefore we test whether the targeted mRNA is carried outside by adding a loading sequence. Third, we test whether viral remnants lacking their natural genome but instead possess the targeted mRNA are able to infect other E.coli and whether we can detect translation. In this assay, we select the Cre generator and the Gfp generator as the targeted mRNA. If we confirm these three work, we can say our MS2 phage system functions as Signal Virus.
1. Titer test of wild type MS2 phage
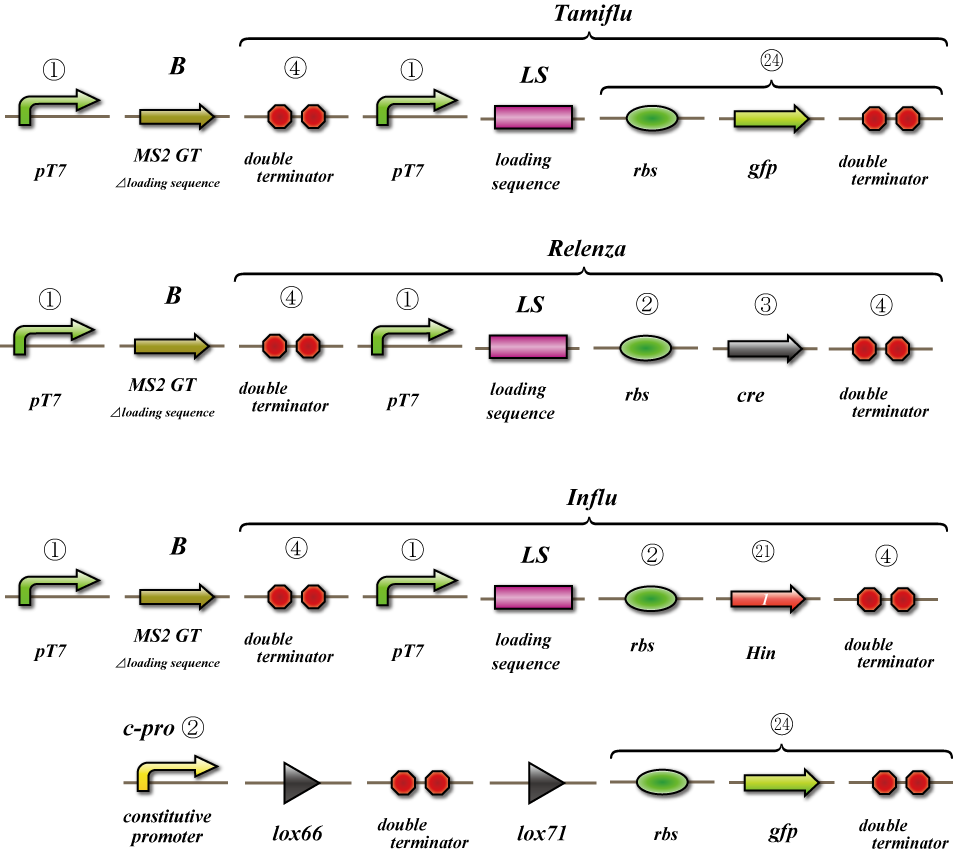
In this experiment, we test whether cDNA of MS2 phage genome produces functional viruses. We transform the plasmid coding for the MS2 phage genome into DE3, and induce its expression by T7 RNA polymerase. After that we collect the cell lysate and perform a titer test by infecting JM109 (Figure 1).
2. Measurement of quantity of the targeted mRNA
In this experiment we test whether the targeted mRNA is packaged by MS2 coat proteins and carried out of the cell. We use the construct shown in Figure 2. We transform the plasmids coding MS2 phage genome to DE3 cells and induce expression using the T7 polymerase. At the same time we induce transcription from the Gfp generator or The Cre generator each with a loading sequence attached. Then the mRNA is packaged and carried out of the cell. After that we collect the cell lysate and the viral remnants it contains. Next we measure the quantity of mRNA coding for the gfp generator or cre generator. It is possible that the detected mRNA is derived from the cytosol of E. coli instead of that derived from the virus. In order to quantify cytosolic RNA, we use as control E.coli transformed with cDNA of MS2 phage and the cre generator or gfp generator and quantify the mRNA coding for the cre generator and gfp generator using the same methods. The difference between the samples and the control represents the quantity of mRNA carried out from the cells because mRNA detected under this condition should be derived from the cytosol. We expect the detection of mRNA carried outside by the viral remnants.
3. Expression test in receiver E.coli
In this experiment we test whether viral remnants lacking their natural genome but instead possess the targeted mRNA are able to infect other E.coli and whether we can detect translation. We use the construct shown in Figure 3. We transform the plasmids coding MS2 phage genom e to DE3 cells and induce expression using the T7 polymerase. At the same time we induce transcription from the Gfp generator or The Cre generator each with a loading sequence attached. Then the mRNA is packaged and carried out of the cell. After that we collect the cell lysate and the viral remnants it contains. And we make concentrated phage solution. We test whether we can detect Gfp is translated in the infected E.coli. On the other hand as the receiver, we prepare E.coli which can express gfp because the terminator is removed by Cre, and infect phage to it. Although gfp can be expressed on both systems, the intensity of gfp emission will not be detected at the same time because in many cases just a few phages can be infected by E.coli. When the gfp expression is detected on either one of the systems, we can conclude that phage in the transmitter E.coli carry out target mRNA and infect it to other bacteria, and in the result, target mRNA is translated in the infected E.coli. The construct of Hin is prepared for negative-control for gfp generator and cre generator.
 "
"