Team:WashU/Notebook/MolecularBiology
From 2010.igem.org
(Difference between revisions)
(→2010/08/19) |
(→Week of 8/23) |
||
| Line 587: | Line 587: | ||
<br> | <br> | ||
| - | == | + | ==2010/08/26== |
| - | + | *Results from transformation on 8/25 | |
| - | Results from transformation on 8/25 | + | |
Number Plasmid Plate Observations | Number Plasmid Plate Observations | ||
1 pSB1AT3 Tet Lawn of colonies with sprinkled red transformants | 1 pSB1AT3 Tet Lawn of colonies with sprinkled red transformants | ||
| Line 597: | Line 596: | ||
5 pSB1K3 Kan ~80 colonies observed all red transformants | 5 pSB1K3 Kan ~80 colonies observed all red transformants | ||
6 None Kan No colonies observed | 6 None Kan No colonies observed | ||
| + | <br> | ||
Latest revision as of 06:52, 27 October 2010

2010/07/01
- Plated The Megax BH10B strain and Strain 8 from the cohen lab onto an amp plate to check that the Megax BH10B strain is killed
2010/07/13
- Minipreped all the cultures and nanodropped them
- Ran 3 PCR reactions (@ 58,60, and 62 C) with primers p1 and p2 in order to attach the kozak onto the YFP
- Enzyme digested Nat (bglII and EcoRI), Kan (BamHI and EcoRI), and Promoter (EcoRI). Did 2 reactions each with ~500 ng DNA
2010/07/14
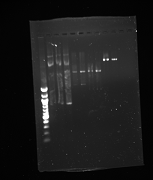
- Gel from July 13th
- Lanes from top to bottom:
1) Ladder 2) 5uL of 58 degree YFP + 1uL of loading dye 3) 5uL of 60 degree YFP + 1uL of loading dye 4) 5uL of 62 degree YFP + 1uL of loading dye 5) 5uL of KanMx4 1 + 1uL of loading dye 6) 5uL of KanMx4 2+ 1uL of loading dye 7) 5uL of NatMx4 1 + 1uL of loading dye 8) 5uL of NatMx4 2+ 1uL of loading dye 9) 5uL of Promoter 1 + 1uL of loading dye 10) 5uL of Promoter 2 + 1uL of loading dye
2010/07/15
- Gel reads from left to right as follows:
Dna Ladder; NatMX4-Promoter construct 2x; KanMX4 plasmid backbone 2x.
- DNA ladder information is here at http://www.neb.com/nebecomm/products/productN3232.asp
2010/07/16
- Transformed EColi with the Kan plasmid containing the NatMX4 Promoter sequence.
- Bacteria Transformation July 16, 2010
1) Thaw competent cells on ice right before transformation. 2) Make 3 100uL tubes of competent cells 3) Add 4 ul of plasmid to cells, flick gently. 4) Incubate on ice 30 min. 5) Heat-shock cells for 30 seconds at 42oC w/o shaking 6) Transfer to ice, incubate 2 min 7) add 250 ul room temp SOC Media 8) Shake @ 37o for 1 hr 9) Spread 150 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10) Incubate o/n at 37oC upside down
2010/07/19
- Transformation of NatMX4/Promoter vector failed.
- Analysis of gel purification showed that the separate pieces which were ligated together on Thursday July 15th were of the correct length.
- Conclusion: the ligation failed; most likely due to denatured ligase. The ligase icebath had melted prematurely and heat inactivated the enzyme.
- Yeast group obtained several replacement ligases and necessary buffers from the Cohen Laboratory in the 4444 building of the WashU Med School.
- Transformation of C2 and SxL constructs into E. coli (AH)
- Reconstituted 5ug of each construct (in plasmid) in 10uL dH20. Final conc. of this stock solution is 500ng/uL. 10uL of working stock of each solution was made by diluting stock solution 1:10. Final conc. of working stock is 50ng/uL.
- Thaw 300uL tube of DH5alpha competent cells and divide among 5 tubes (60uL each). Amounts of 50ng/uL working stock were added to tubes labeled A-E in the following way:
A: 1uL C2 B: 5uL C2 C: 1uL SxL D: 5uL SxL E: Control=no DNA
- Incubate on ice 30 min, heat shock for 30s at 42C, rest on ice for 2 min.
- Add 250uL of LB to each tube, Shake at 37C for 1hr
- Plate 150uL on warm LB + Amp plates o/n at 37C
2010/07/20
- Digested 4 sequences:
1) NatMx4 with BglII and EcoRI: in microliters [5 buffer 3(NEB); 2.5 DNA at 200 ng/uL; 0.5 of BSA; 1 of each enzyme; 40 of H2O.] 2) Constitutive Promoter with EcoRI and SpeI: in uL [5 buffer 4(NEB); 2.5 DNA at 200 ng/uL; 0.5 of BSA; 1 of each enzyme; 40 of H2O.] 3) Construct 2 with BglII and XbaI: in uL [5 buffer 2(NEB); 1 DNA at 500 ng/uL; 0.5 of BSA; 1 of each enzyme; 41.5 of H2O.] 4) SxL with BamHI: in uL [5 buffer 3(NEB); 1 DNA at 500 ng/uL; 0.5 of BSA; 1 of BamHI; 41.5 of H2O.]
- all were incubated for 4 hours followed 20 minutes of heat inactivation.
2010/07/21
- Digest SxL construct with EcoRI: added 1uL EcoRI to already done mix from 20 July digestion.
- Gel purify NatMx4, Promoter, Construct 2, and KanMx4 digest results from 20 July 2010
Note: KanMx4 bands were not observed but approximate region of gel was removed for ligation KSB.
- Ligate all three pieces together: NatMx4, Promoter, and Construct 2
- 2 reactions:
1) NCPA = NatMx4+Promoter+Construct 2
3.5uL Nat + 3.5uL Pro + 1.5uL C2
2) NCPB = NatMx4&Promoter+Construct 2
[NatMx4&Promoter was pre-existing ligation product from 15/16 Jul]
7uL NP + 1.5uL C2
- Ligate KanMx4 with Sxl
- 2 reactions:
1) KSA = KanMx4 Digest 2 + SxL
7uL KanDigest2 + 1.5uL SxL
2) KSB = KanMx4 (purified) + SxL
7uL GelPureKanMx4 + 1.5uL SxL
- Freezer stocks of C2 and SxL
- 1:1 ratio of LB was mixed with 100% glycerol to make a 50% glycerol/LB solution.
- 750uL of glycerol/LB solution was added to 750uL of each C2 and SxL culture from transformation on 2010/07/19.
- Two tubes of each were placed at -80C.
2010/07/22
- Transformed the ligation DNA from reactions NCPA, NCPB, KSA, and KSB into E. Coli using the following protocol.
- Bacteria Transformation
1. Thaw competent cells on ice right before transformation. 2. Add 4ul of plasmid to cells, flick gently. 3. Incubate on ice 30 min. 4. Heat-shock cells for 30 seconds at 42oC w/o shaking 5. Transfer to ice, incubate 2 min 6. add 125 ul room temp LB Media 7. Shake @ 37o for 1 hr + 8. Spread 100 ul on a pre-warmed plate w/ correct antibiotic selector, allow sample to dry on plate 9. Incubate o/n at 37oC upside down
2010/07/26
- Digest and gel purify to confirm the results of the SxL construct transformants created on July 22.
- Gel image is attached:
- Reactions 4, 5, 6, and 7 of the Yeast Group's gel are from July 21 ligation, July 22 transformation, and the July 24 miniprep of the July 23 cultures. [The leftmost lane is a ladder; numbering begins at the second lane with Number 1.]
- Sequencing the YFP to confirm the Kozak Sequence:
- iGEM1 YFP 62 degrees C primer P9
- iGEM2 YFP 62 degrees C primer P2
- iGEM3 YFP 62 degrees C primer P9
- iGEM4 YFP 62 degrees C primer P2
2010/07/27
- Digestion of Terminator and YFP pieces (YFP is currently being sequenced to confirm Kozak Sequence)
- Terminator
• 37.5 uL of H20 • 5 uL of NEB buffer 3 • 5 uL of Terminator DNA • 1 uL of Pst1 enzyme • 1 uL of Xba1 enzyme • 0.5 uL of BSA
- YFP
• 37.5 uL of H20 • 5 uL of NEB buffer 2 • 5 uL of YFP DNA • 1 uL of Pst1 enzyme • 1 uL of Spe1 enzyme • 0.5 uL of BSA
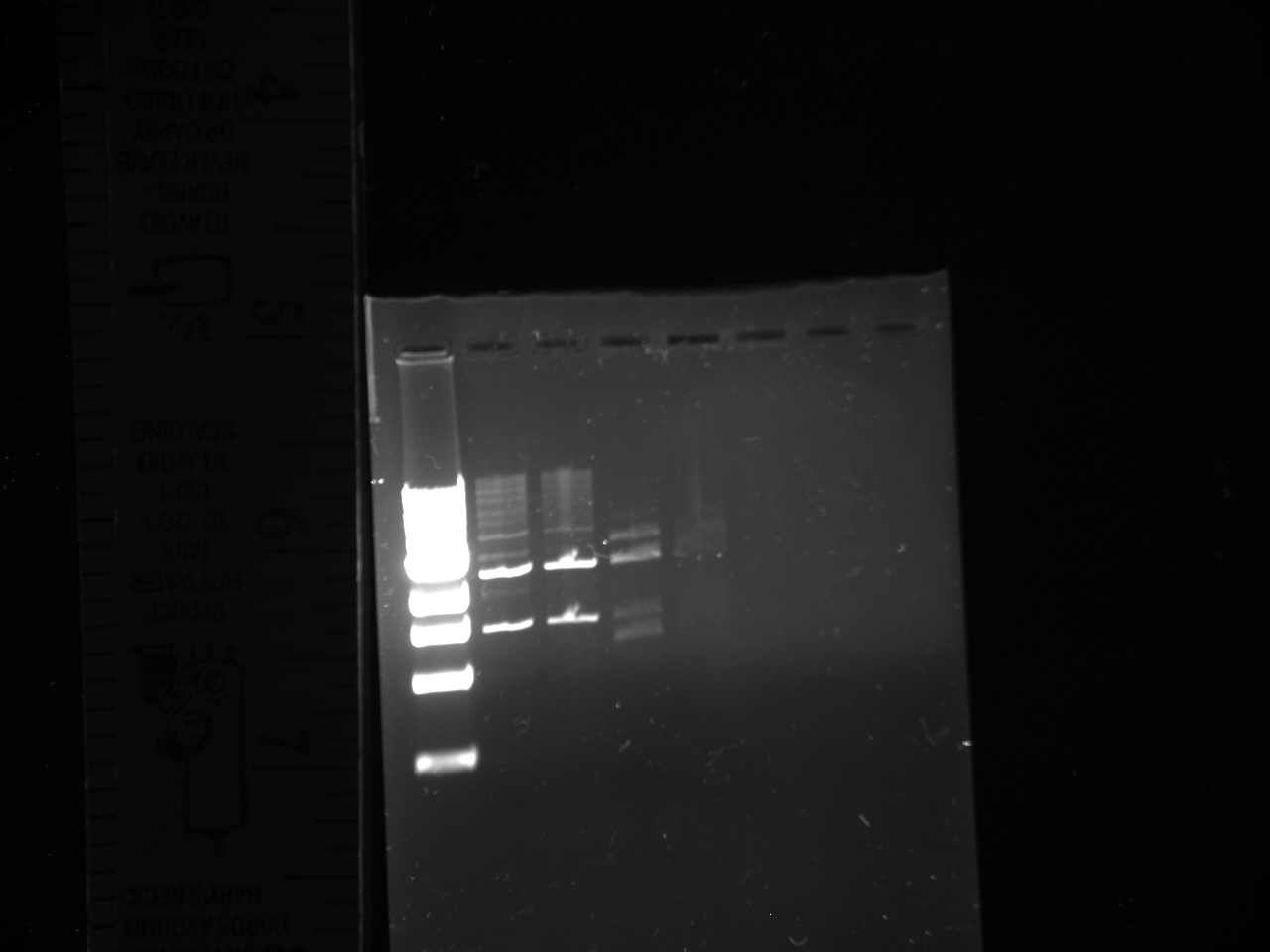
- Gel Purification and Confirmation of NPCA, NPCB, Terminator, and YFP
- Gel Lanes:
1) Ladder 2) NPCA 1 3) NPCA 2 4) NPCB 1 5) NPCB 2 6) Terminator 7) YFP 8) YFP
2010/07/28
- Double Digests of KSA2 (confirmed); NPCA 1 & 2; and NPCB 1 & 2.
• 32.5 uL of H2O • 10 uL of DNA • 5 uL of NEB buffer 4 • 1 uL of XhoI • 1 uL of AvrII • 0.5 uL of BSA
- Began Colony PCR of NPCA and NPCB plates: 14 colonies taken from each.
2010/07/29
- Colony PCR:
- Primers p1 and p5 are defective Colony PCRs will not yield proper results
- The back up cultures still are intact and can be reused when the proper primers arrive
- Digestions:
- YFP Biobrick:
• 32.5 uL of H2O • 5 uL of DNA • 5 uL NEB buffer 3 • 5 ul CIP • 1 uL PstI • 1 uL XbaI • 0.5 uL BSA
- YFP kozak [Mix is defective due to Primer 1 error; DNA does not contain XbaI site]
• 30 uL kYFP DNA • 12.5 uL of H2O • 5 uL NEB buffer 4 • 1 uL XbaI • 1 uL SpeI • 0.5 uL BSA
- C2 control
• 37.5 uL H2O • 5 uL DNA • 5 uL NEB buffer 4 • 1 uL XhoI • 1 uL AvrII • 0.5 uL BSA
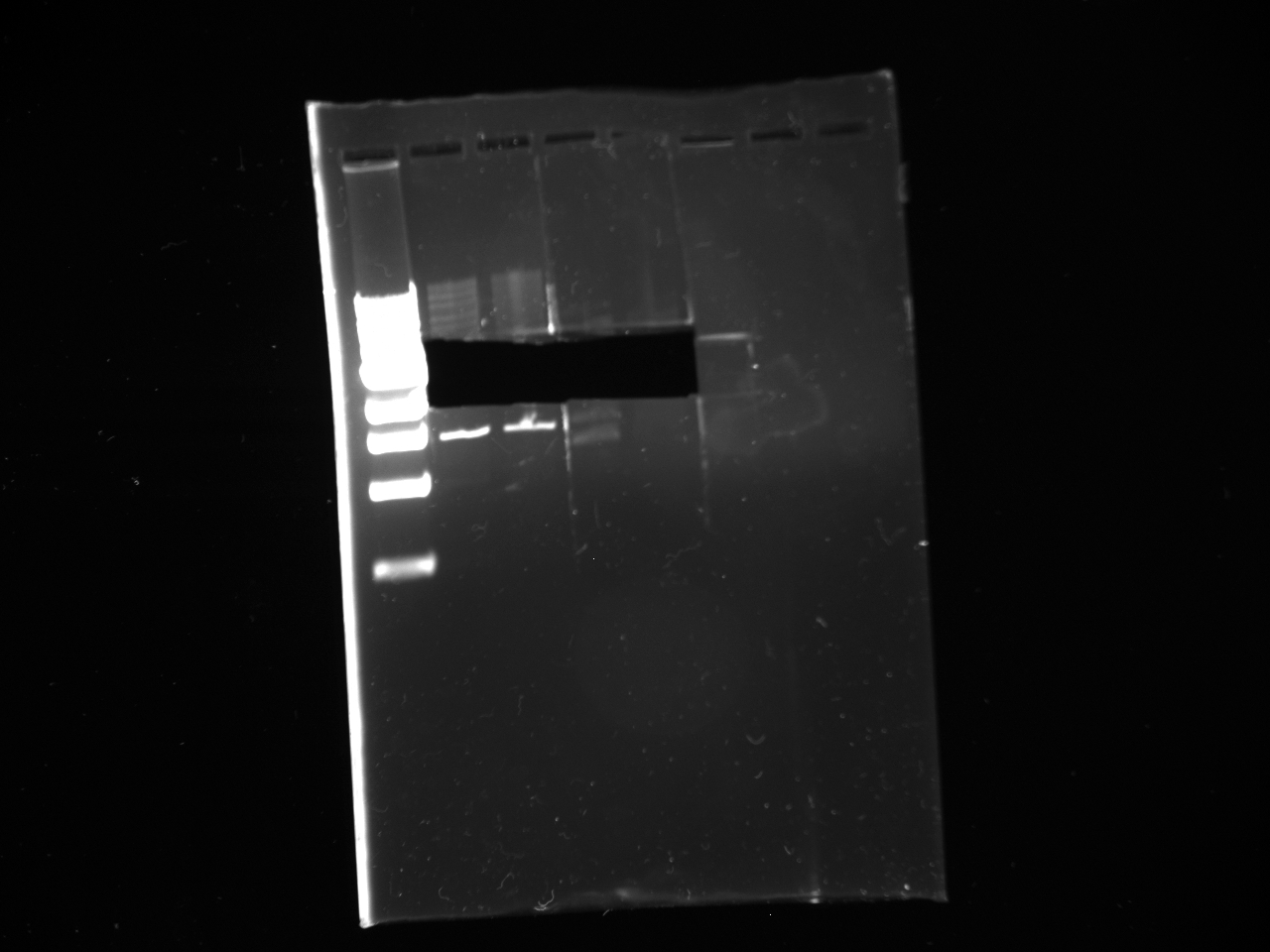
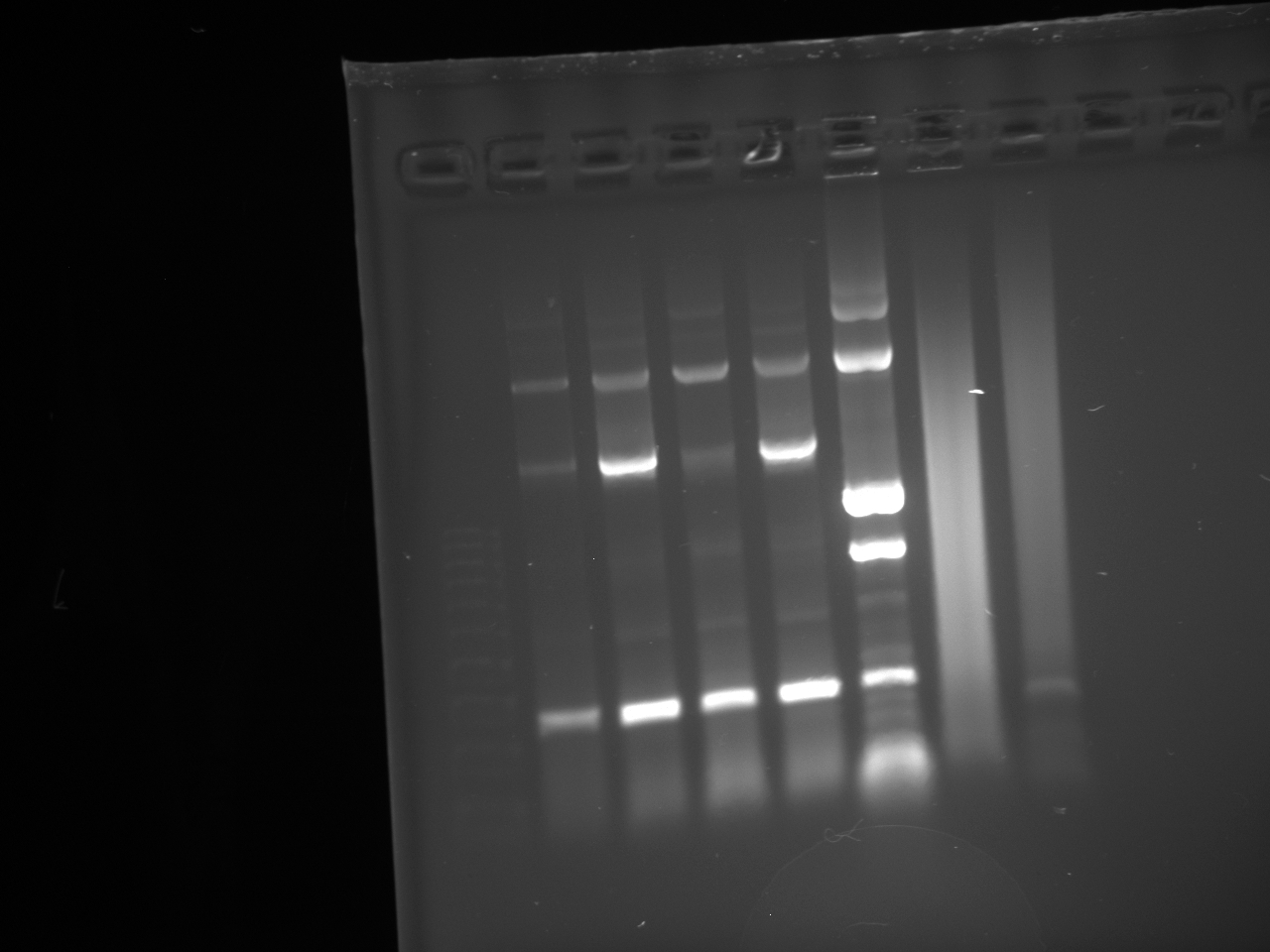
- Gel:
• Lane 1: Ladder 1kb • Lane 2: C2 control • Lane 3: NPCA1 3.7 and 2.3kb • Lane 4: NPCA2 3.7 and 2.3kb • Lane 5: NPCb1 3.7 and 2.3kb • Lane 6: NPCb2 3.7 and 2.3k
- Conclusion NP was not present within the construct 2 plasmid
- Gel 2:
- YFP backbone--- not observed Gel tossed out
- Ligations:
- NPCc: ligate C2// BglII & XbaI with CIP A with NatMx4 and Promoter
• 1uL Y4 ligase buffer • 0.5 uL ligase • 3 uL NatMx4 • 3 uL Promoter • 2.5 uL C2
- NPb: NatMX4 ligates to Promoter
• 17 uL NatMx4 • 17 uL Promoter • 4 uL T4 ligase buffer • 2 uL ligase
- Extra Digestion:
- C2 // bglII & XbaI CIP B
• 27.5 uL H2O • 10 uL DNA • 5 uL CIP • 5 uL NEB buffer 3 • 1 uL BglII • 1 uL XbaI • 0.5 uL BSA
2010/07/30
- Gel purify 30 uL NPb and 30 uL C2b
- Make NPCd: Ligate NPb & C2b (non purified) ~ 10 uL of each
• 5.5 uL NPb • 3 uL C2b • 0.5 ligase • 1 uL buffer
- Make NPCe: Ligate NPb & C2b (purified)
• 24 uL of NPb DNA • 1.5 uL of C2b DNA • 3 uL buffer • 1.5 uL ligase
- Re-attempt ligation of NatMx4 and Promoter
• 25.5 uL mix of NatMx4 and Promoter • 3 uL buffer • 1.5 uL ligase
2010/08/02
- Transformations: NPCc NPCd NPCe and KSA2
1. Thaw competent E.Coli cells on ice right before transformation. 2. Make 4 tubes with 70uL of competent cells each 3. Add 3 ul of NPCc, d and e plasmid to 3 tubes and 1 uL of KSA2 to the last tube, flick gently. 4. Incubate on ice 30 min. 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 140 ul room temp LB Media 8. Shake @ 37o for 2 hr [to ensure recovery from shock] 9. Spread 100 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
- PCR reactions:
- Reaction 1st round
kYFPa: 94ng of YFP at 59 degrees C using p10 & p2 kYFPb: 94ng of YFP at 61 degrees C using p10 & p2
- Reaction 2nd Round
kYFPa1: 1 uL of kYFPa at 59 degrees C using p11 & p2 kYFPb1: 1 uL of kYFPb at 59 degrees C using p11 & p2 kYFPa2: 1 uL of kYFPa at 61 degrees C using p11 & p2 kYFPb2: 1 uL of kYFPb at 61 degrees C using p11 & p2
2010/08/03
- Cultures:
- Created liquid cultures from the transformation results of Ksa2, NPCc, NPCd, NPCe, and C2.
[C2 transformation was performed by Amanda when the C2 construct arrived]
- Digests: kYFPa1, a2, b1, b2 and YFPBiobrick on backbone pSB1C3
- For ALL:
• 27.5 uL of H2O • 15 uL of DNA of the respective construct • 5 uL of NEB buffer 3 • 1 uL of PstI enzyme • 1 uL of XbaI enzyme • 0.5 uL of BSA
- [resulting products were column purified and eluted to 50 uL]
- Ligation of kYFP's and the backbone
• 21 uL of kYFPa1, a2, b1, b2 in 4 respective tubes • 4.5 uL of BB into all four tubes • 3 uL buffer • 1.5 uL T4 ligase
- Of NatMx4 and Promoter
- NatMx4
• 35 uL of H2O • 7.5 uL of DNA • 5 uL of NEB buffer 3 • 1 uL of bglII • 1 uL of EcoRI • 0.5 uL BSA
- Promoter
• 35 uL of H2O • 7.5 uL of DNA • 5 uL of NEB buffer 4 • 1 uL of EcoRI • 1 ul of SpeI • 0.5 uL BSA
2010/08/04
- Gel purification of NatMX4 and Promoter
- 4 lanes Nat and 4 lanes Pro
- Successfully extracted the lower of 2 visible lines from all lanes [top line was vector; bottom was Nat and Pro inserts]
- Miniprep: Kan-SxL construct and Construct 2
- Transformation: kYFPa1, a2, b1, b2 on the Biobrick Backbone.
1. Thaw competent cells on ice right before transformation. 2. Make 4 75uL tubes of competent cells 3. Add 4 ul of plasmid to cells, flick gently. 4. Incubate on ice 30 min. 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 150 ul room temp LB Media 8. Shake @ 37o for 1 hr 9. Spread 100 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
- Ligation: NatMx4 + Promoter
• 30 uL of DNA (mix of both parts; they were extracted together) • 4 uL of Buffer • 4 uL of H2O • 2 uL of T4 ligase
2010/08/06
- Transformation: kYFP on the Biobrick Backbone.
1. Thaw competent cells on ice right before transformation. 2. Make 1 100 uL tubes of competent cells 3. Add 4 ul of plasmid to cells, flick gently. 4. Incubate on ice 30 min. (10:45 to 11:15) 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add ul room temp SOC Media 8. Shake @ 37o for 1 hr 9. Spread 100 ul on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
2010/08/09
- Make Tetracycline plates to select for kYFP transformants
- Transform kYFP into E.Coli
1. Thaw competent cells on ice right before transformation. 2. Make a 75 uL tube of competent cells 3. Add 2 ul of plasmid to cells, flick gently. 4. Incubate on ice 30 min. (12:10 to 12:40) 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 125 ul room temp SOC Media 8. Shake @ 37o for 1 hr 9. Spread 100 ul on a pre-warmed tetracycline selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
2010/08/10
- Made a culture from possible colonies on tetracycline plate
- Double Digest of pSB1A3
• H2O • DNA • NEB buffer 3 • XbaI • PstI • BSA
- Gel purification of digest products
- 1kb ladder, 3 lanes of pSB1A3, and 3 lanes of YFP-BB
- results: all 3 bands observed (one for the pSB1A3 digest and 2 for the YFPbb) [an extra band from the biobrick was seen above the 2 desired YFP bands possibly single or uncut plasmid]
- Gel extraction of digest products
- 1 extraction for each of the 3 desired bands
- Ligations
• kYFP to pSB1A3 • kYFP to Biobrick backbone • YFP to pSB1A3 • YFP to Biobrick backbone
- Transformation of the ligation products into E. Coli
1. Thaw competent cells on ice right before transformation. 2. Make 4, 70 uL tube of competent cells 3. Add 2 l of plasmid to cells, flick gently. 4. Incubate on ice 30 min. (3:45 to 4:15) 5. Heat-shock cells for 30 seconds at 42oC w/o shaking 6. Transfer to ice, incubate 2 min 7. add 125 l room temp SOC Media 8. Shake @ 37o for 1 hr 9. Spread 100 l on a pre-warmed ampicilin selector plates, allow sample to dry on plate 10. Incubate o/n at 37oC upside down
2010/08/11
- Colony PCR
• 4 possible transformant colonies PCR'd and back-up culture made
- PCR
• Re-PCRing the kYFP at 59, 60, 62 degrees C [product was named kYFP2]
- Transformation of kYFP with o/n ligation results
• 50uL of cells • 2 uL of kYFP on Biobrick backbone • 2 uL of kYFP on pSB1A3 • 150uL of SOC media • plated all of the incubated/shaken culture
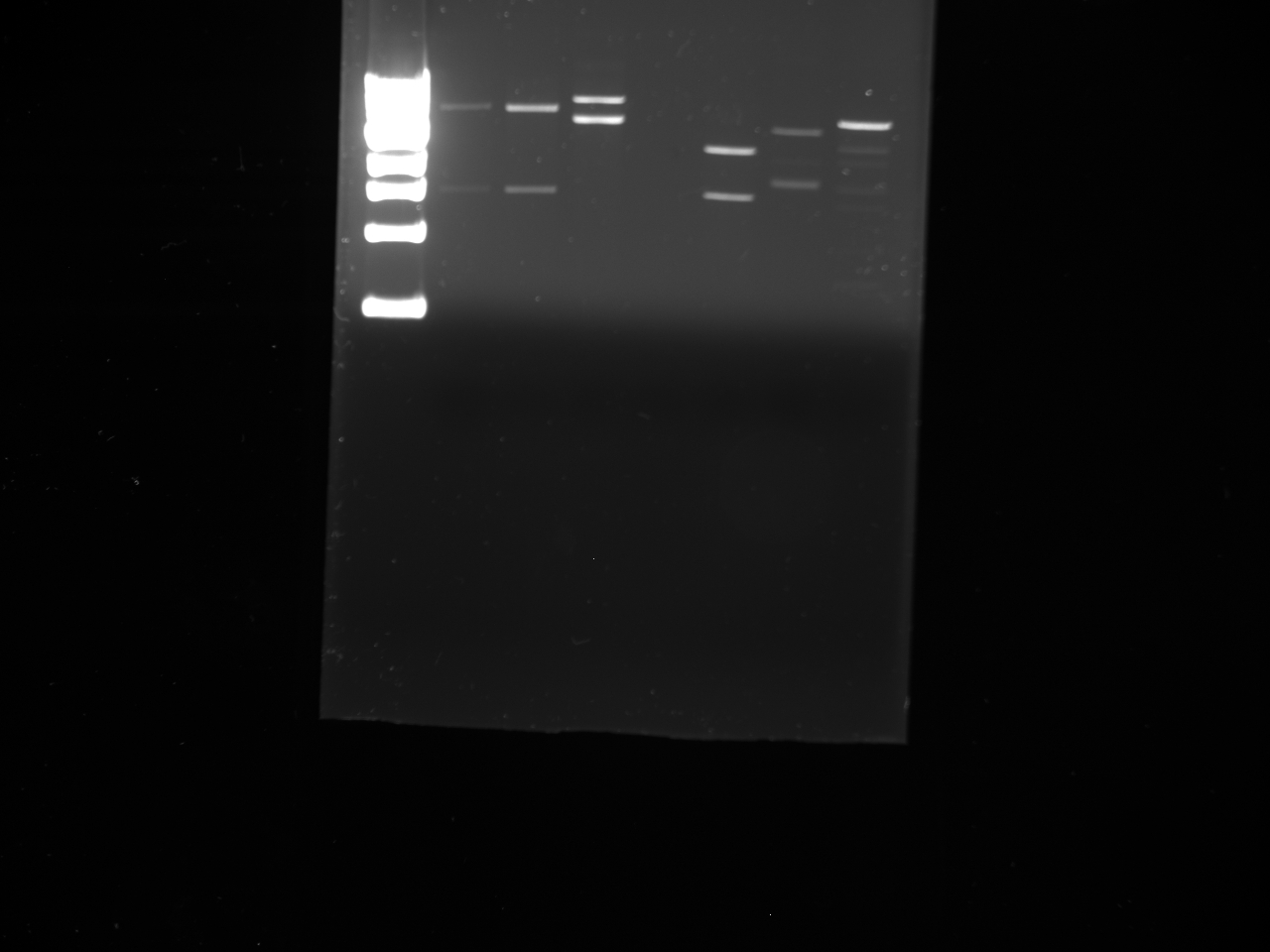
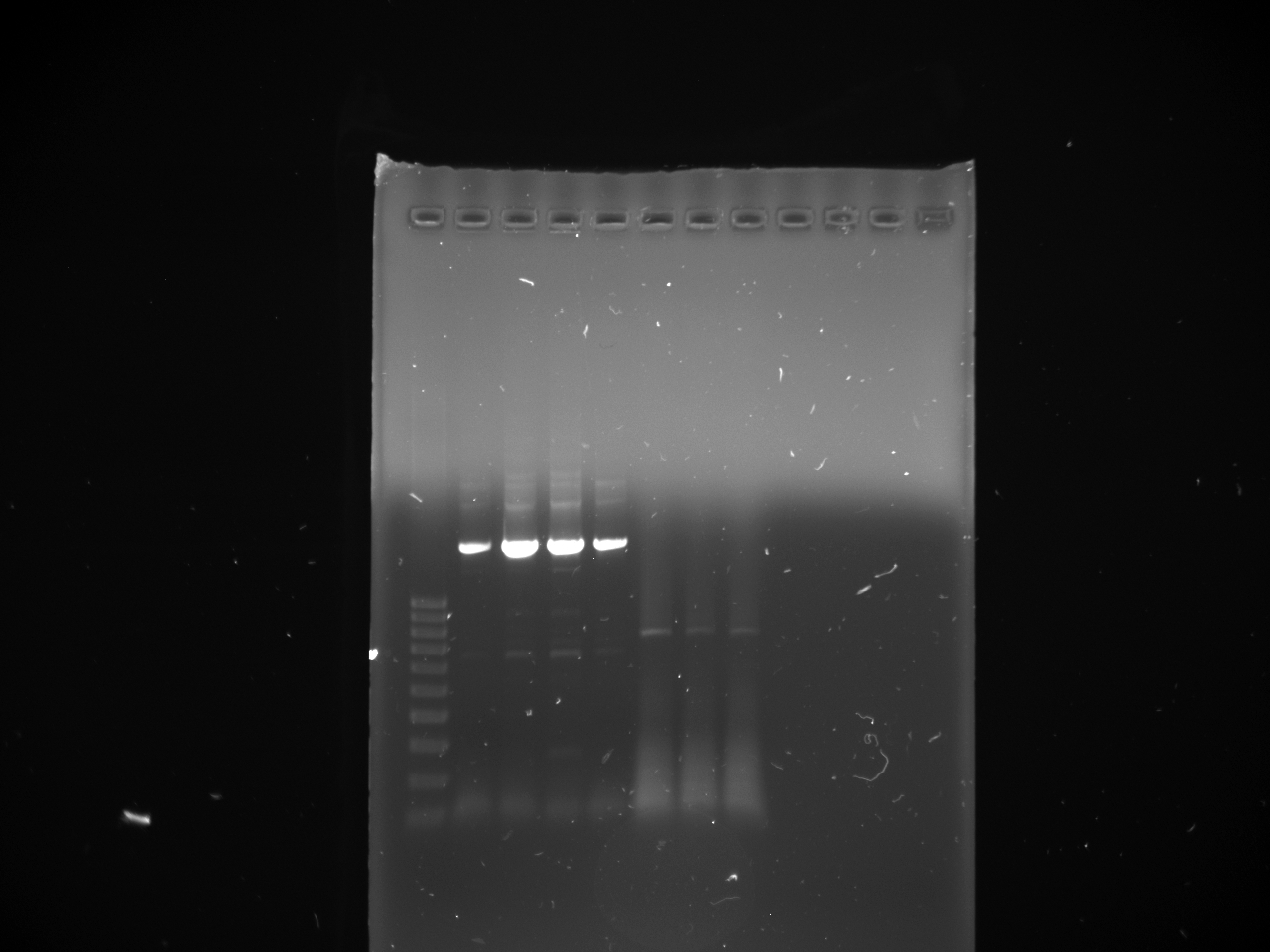
- Gel purifications
- 100b ladder; 4 lanes of colony PCR; 59*C PCR of new kYFP; 60*C PCR of new kYFP; 62*C PCR of new kYFP.
- results:
- Digests of PCR Products and the Terminator
- Digesting PCR kYFP with XbaI and SpeI
- Digesting terminator [in vector] with XbaI and CIP
- Ligation
2010/08/12
- Miniprepped the cultures made on the 11th [#4 had no growth or pellet]
- Created cultures of the 4 transformant colonies from the 11th's transformation
- Redesigned the kYFP reverse primer
- Ligate Terminator and kYFP2 (from the 11th's digestion)
- [30min room temp ligation]
- PCR at 60*C the Miniprep results, a YFP control and the Ligation product kYFP2term with primers p2 and p12& p9 and p12 reactions with the YFP control and the kYPF2term. [7 rxns total as #4 yielded no pellet for miniprepping]
- [p2 was the questionable reverse kYFP primer]
- Gel run: 100bp ladder, culture 1 DNA; cult. 2 DNA; cult. 3 DNA; YFP 2-12 PCR; YFP 9-12 PCR; kYFP2 2-12 PCR; kYFP2 9-12 PCR
- Results
- Conclusion: Colonies were not desired kYFP transformants [match the pattern of the YFP (2-12) control]
- Transformation of E.Coli with the kYFP2term plasmid (ligation product)
3uL in 50uL of cells 150uL SOC media 75 ul plated onto amp selector plates
2010/08/13
- Transformation yielded a large number of colonies which may contain the kYFP2term construct
- Pour new ampicilin plates
- Beginning screening processes today:
- Grow 16 cultures of several colonies from transformants
- Future screening steps:
- Miniprep the cultures
- Digest DNA with XbaI & PstI
- Digest C2 with XbaI & PstI
- Gel12 purify C2 and the correct kYFP2term from the improper kYFP2term [X will cut improper into 3 pieces, correct into 2]
- 30min room temp ligate correct kYFP2term with C2
- Transform the ligated construct, C2kYFP2term, into E. Coli
- Grow cultures from transformation
- Miniprep the cultures
- Submit DNA for sequencing
- Digest with bglII & XbaI & CIP
- Gel purify
- 30 min room temp ligation of C2kYFP2term with the NatMX4-Promoter construct
- Transform ligation product into E.Coli
2010/08/14
- Miniprepped the cultures (16) made from kYFP2term transformants
- Digests:
- Digested C2 with SpeI and PstI and CIP in NEBuffer 2. [27.5 H2O; 14 DNA 1 CIP; 1 SpeI; 1 PstI; 5 buffer; 0.5 BSA]
- 6 Digests of Miniprepped DNA with XbaI and PstI in NEBuffer 2 [35.5 H2O; 7 DNA; 5 buffer; 1 XbaI; 1 PstI, 0.5 BSA]
Note: rxns 12,13,14,15, and 16 used low concentration XbaI
- Gels:
- 12 lane gel contained: 1kb ladder; culture samples 1-11
- 8 lane gel contained: 1 kb ladder; culture samples 12-16
- Expected results
1. Successful construction: 2 kb band and .993kb band 2. Backwards kYFP2: 3 kb band 3. No kYFP2: 2kb band and .25 kb band
- Results
- Conclusion: #3 results for all samples; no kYFP2 ligated into the terminator
2010/08/19
- Got the new primers in today.
- Ran a PCR to make the new KYFP inserts.
- Used 1 ul of YFP biobrick plasmid as the template.
- p15 and p 16 were used to make the KYFP3.
- p10 and p17 were used to make the preKYFP4.
- Three PCR reactions were set up for each and were then run at 58, 60, and 64 C with 1 min extension time.
- The PCR products were then run out on a gel.
- The bands coresponding to ~ 800 bp were cut out and gel purified.
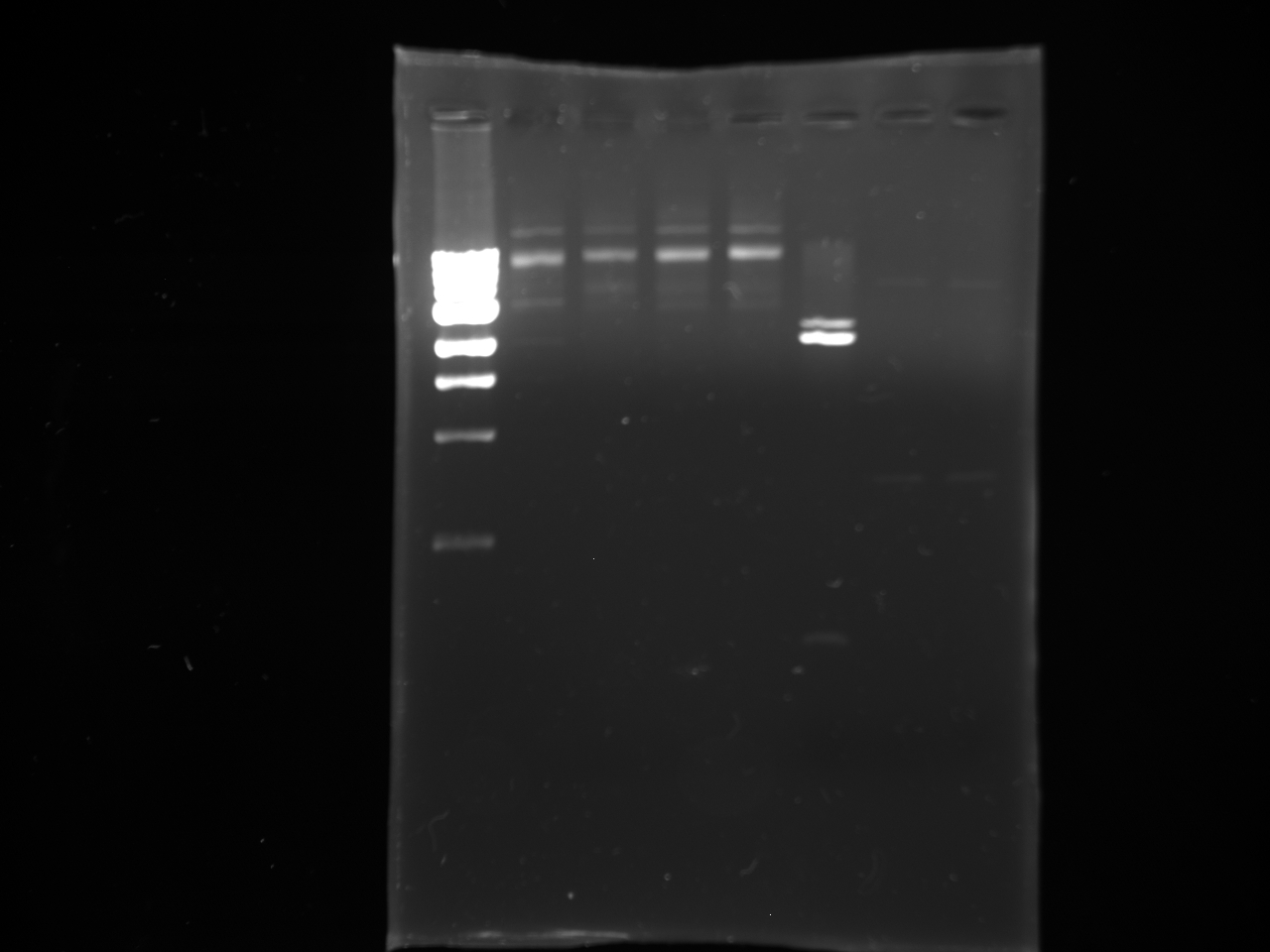
- Lanes:
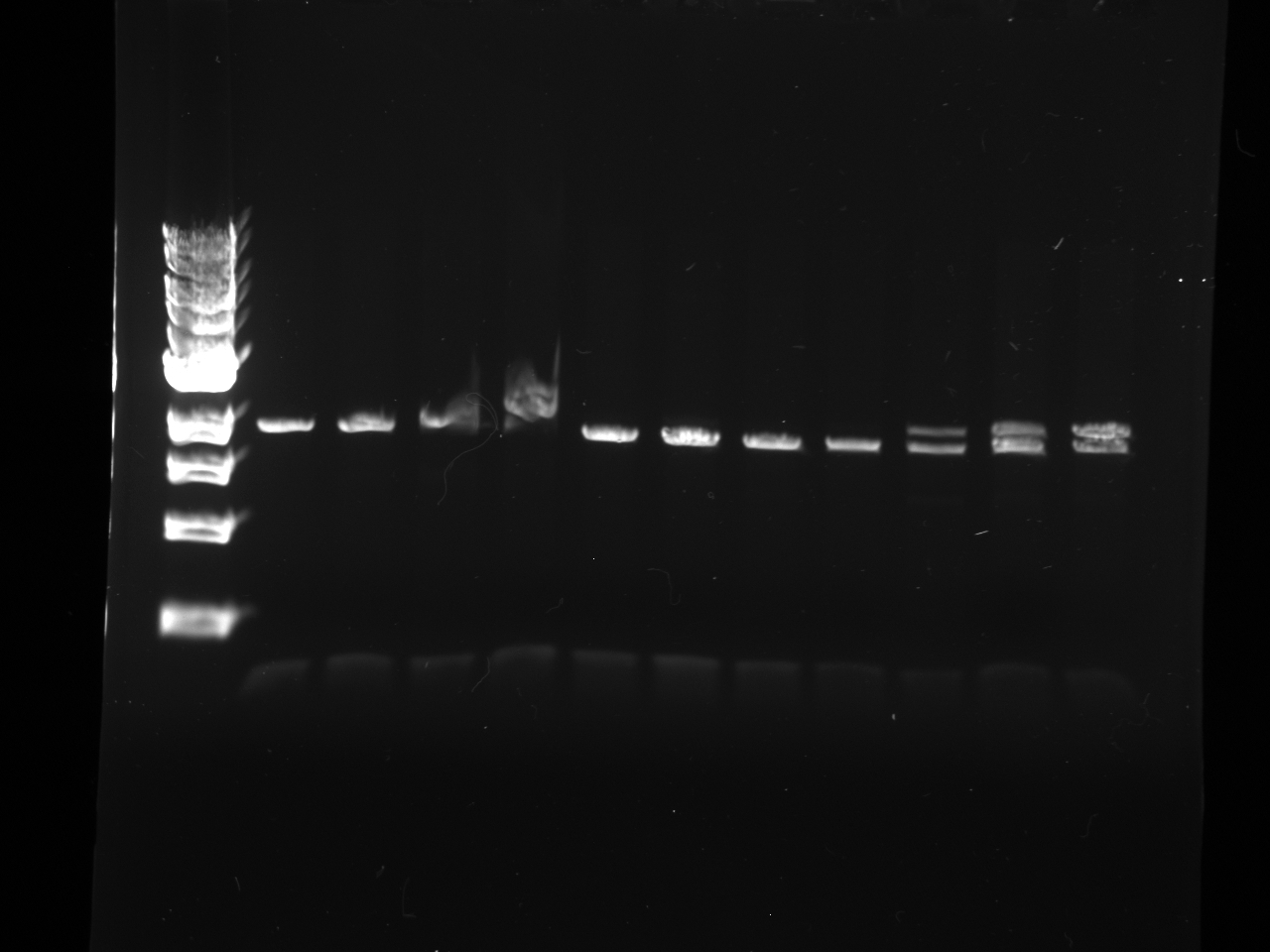
1 100 bp ladder 2,3 KYFP3 @58 4,5 KYFP3 @60 6,7 KYFP3 @64
- Lanes:
1 100 bp ladder 2,3 preKYFP4 @58 4,5 preKYFP4 @60 6,7 preKYFP4 @64 8 100 bp ladder
- The preKYFP4 products were cleaner as was to be expected.
- The KYFP3 @60 has anomalous lower bands ~200 bp and the KYFP3's in general have more minor banding patterns
- Running a PCR over night.
• 1 ul of KFYP3 64+p13+p16=KYFP5 o running 2 reactions: one at 60 and the other at 62 C • 1 ul of preKYFP4 64+p11+p16=KYFP4 o running 2 reactions: one at 60 and the other at 62 C
2010/08/20
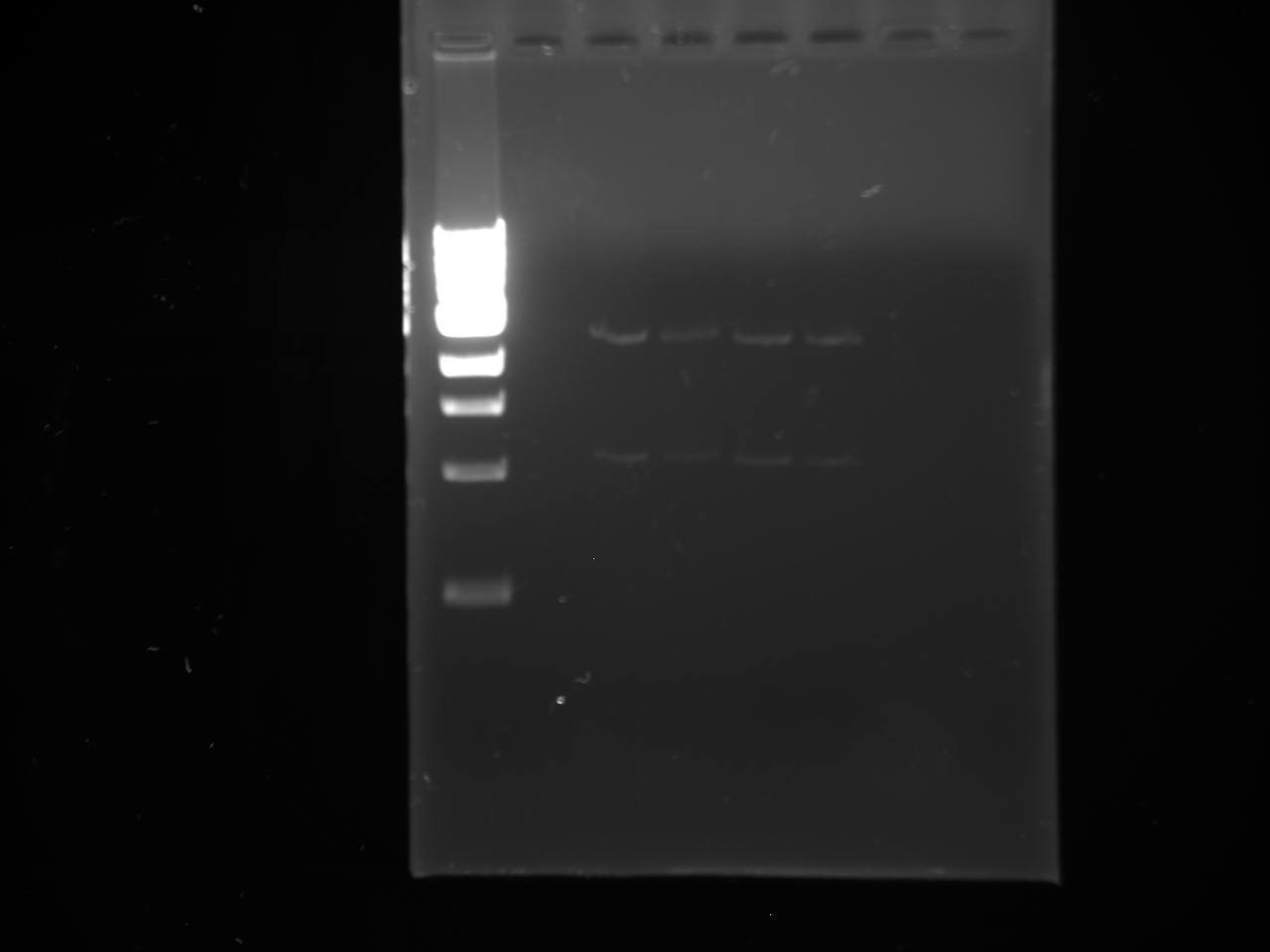
- Ran the PCR products out on a gel.
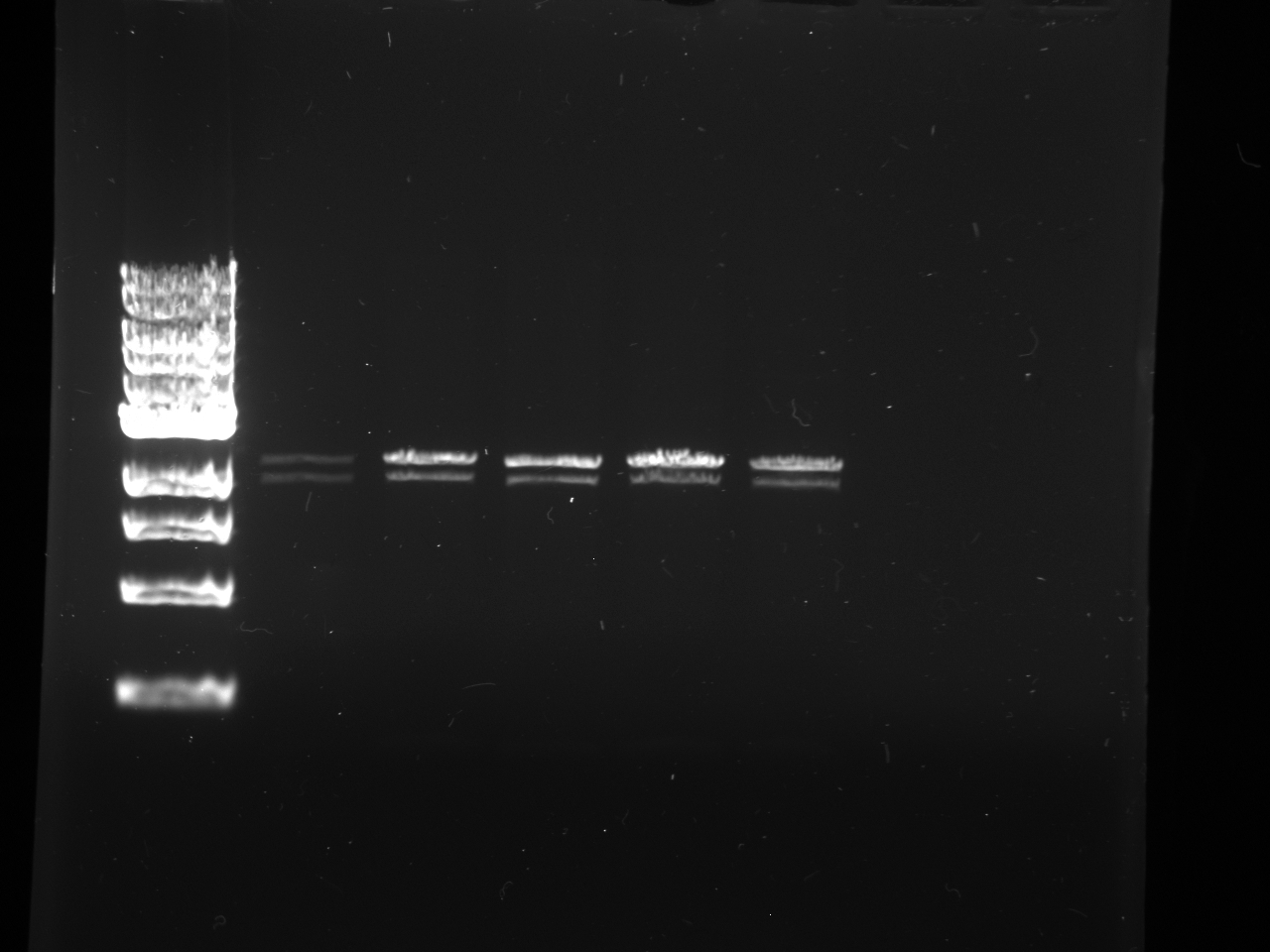
- Lanes:
1 100 bp ladder 2 KYFP5 @60 3,4 KYFP4 @60 5,6 KYFP5 @62 7,8 KYFP4 @62
- The bands that are ~800 bp were cut out and purified.
- Then I set up a five hour enzyme digest @ 37 C.
- I cut KYFP3 @64 w/ XbaI and PstI, KYFP4 @62 w/ XbaI and PstI, KYFP5 @62 w/ XbaI and PstI, YFP backbone w/ XbaI, PstI, and CIP, and Terminator with XbaI and CIP.
- The digested results were run out on a gel and then the appropriate bands were excised and the DNA was purified out.
- An over night RT ligation was set up with 1 ul of backbone: 7.5 ul insert.
- Cut KYFP3, KYFP4, and KYFP5 were ligated into the cut YFP back bone.
- A negative control (only cut YFP back) and a positive control (YFP insert back into YFP back bone) were also set up.
2010/08/26
- Results from transformation on 8/25
Number Plasmid Plate Observations 1 pSB1AT3 Tet Lawn of colonies with sprinkled red transformants 2 None Tet Lawn of colonies 3 pSB1AT3 Amp ~500 colonies, all small, mostly red transformations but some 4 None Amp ~100 colonies, some very large in size 5 pSB1K3 Kan ~80 colonies observed all red transformants 6 None Kan No colonies observed
 "
"