Team:Virginia United/Project
From 2010.igem.org
(Difference between revisions)
(→Optimization of Fluorescent Proteins) |
(→References) |
||
| (5 intermediate revisions not shown) | |||
| Line 110: | Line 110: | ||
Magliery et. al (2004) developed a method for detecting protein binding partners by fusing non-fluorescent fragments of GFP to the two peptides of interest. If the attached peptides sufficiently attract one another, the GFP | Magliery et. al (2004) developed a method for detecting protein binding partners by fusing non-fluorescent fragments of GFP to the two peptides of interest. If the attached peptides sufficiently attract one another, the GFP | ||
fragments will be irreversibly reassemble to form a fluorescing GFP protein. We build on their work by demonstrating how GFP fragment reassembly ("fluorescent complementation") can be used to create AND logic. | fragments will be irreversibly reassemble to form a fluorescing GFP protein. We build on their work by demonstrating how GFP fragment reassembly ("fluorescent complementation") can be used to create AND logic. | ||
| - | |||
| - | |||
| - | |||
===== Hybrid Promoters ===== | ===== Hybrid Promoters ===== | ||
| Line 133: | Line 130: | ||
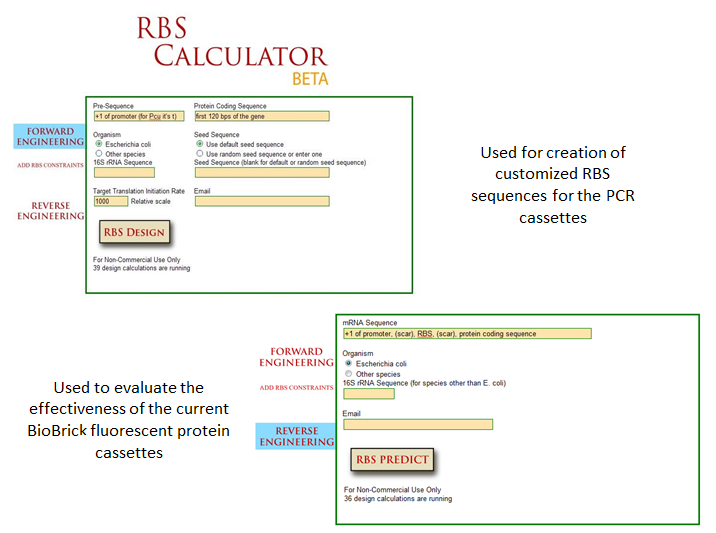
One method of optimization that has recently been explored is the inherent differences in efficiency of gene-specific ribosome binding sites (Voigt et al., 2009). The majority of the time when designing a construct in iGEM, a ‘default’ RBS is used by including just the basic Shine-Dalgarno sequence found in all prokaryotic ribosome binding sites. What many do not realize is that when utilizing the Shine-Delgarno conserved sequence in different gene constructs, the efficiency of the expression for that construct depends on the gene. Utilizing the RBS calculator developed by Voigt et al., one can test the actual optimization rates of the ribosome binding sites by measuring fluorescence output of the final constructs. This allows more control in the system, instituting more flexibility in the precise design of one’s constructs. | One method of optimization that has recently been explored is the inherent differences in efficiency of gene-specific ribosome binding sites (Voigt et al., 2009). The majority of the time when designing a construct in iGEM, a ‘default’ RBS is used by including just the basic Shine-Dalgarno sequence found in all prokaryotic ribosome binding sites. What many do not realize is that when utilizing the Shine-Delgarno conserved sequence in different gene constructs, the efficiency of the expression for that construct depends on the gene. Utilizing the RBS calculator developed by Voigt et al., one can test the actual optimization rates of the ribosome binding sites by measuring fluorescence output of the final constructs. This allows more control in the system, instituting more flexibility in the precise design of one’s constructs. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ==== References ==== | ||
| + | |||
| + | Ball DA, Lux MW, Graef RR, Peterson MW, Valent JD, Dileo J, Peccoud J (2010) Co-design in Synthetic Biology: A System-Level Analysis of the Development of an Environmental Sensing Device. Biocomputing 2010: 385-396. | ||
| + | |||
| + | Balagadde FK, Song H, Ozaki J, Collins C, Barnet M, Arnold FH, Quake SR, You L (2008) A Synthetic Escherichia coli Predator-Prey Ecosystem. Molecular Systems Biology 4: 187 | ||
| + | |||
| + | Collins CH, Arnold FH, Leadbetter JR (2005). Directed Evolution of Vibrio fischeri LuxR for Increased Sensitivity to a Broad Spectrum of Acyl-homoserine Lactones. Molecular Microbiology 55: 712-723. | ||
| + | |||
| + | Collins CH, Leadbetter JR, Arnold FH (2006). Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nature Biotechnology 24: 708-712. | ||
| + | |||
| + | Czar CJ, Anderson C, Bader JS, Peccoud J (2009) Gene Synthesis Demystified. Trends in Biotechnology 10: 63-72. | ||
| + | |||
| + | Fuqua C, Winans S, Greenberg EP (1996). Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol 50: 727-751. | ||
| + | |||
| + | Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339-342. | ||
| + | |||
| + | Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA (2007). USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Research 35: E55. | ||
| + | |||
| + | Magliery et al. Detecting Protein- Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism. J. Am. Chem. Soc (2005) vol. 127 (1) pp. 146-157 | ||
| + | |||
| + | Mukerji S, Oudenaarden A (2009) Synthetic Biology: understanding biological design from synthetic circuits. Nature Reviews Genetics 10: 859-871. | ||
| + | |||
| + | Nasser W, Bouillant ML, Salmond G, Reverchon S (1998). Characterization of the Erwinia chrysnthemi expI – expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Molecular Microbiology 6: 1391-1405. | ||
| + | |||
| + | Salis, H., Mirsky, E.A. and Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression, Nat. Biotechnol. 27, 946 - 950 (2009) | ||
Latest revision as of 21:21, 27 October 2010
 "
"