Team:Stockholm/Project Idea/Proteins
From 2010.igem.org
(Difference between revisions)
m (→Human basic fibroblast growth factor, bFGF) |
m (→IgG protease (IdeS)) |
||
| (19 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
== Proteins == | == Proteins == | ||
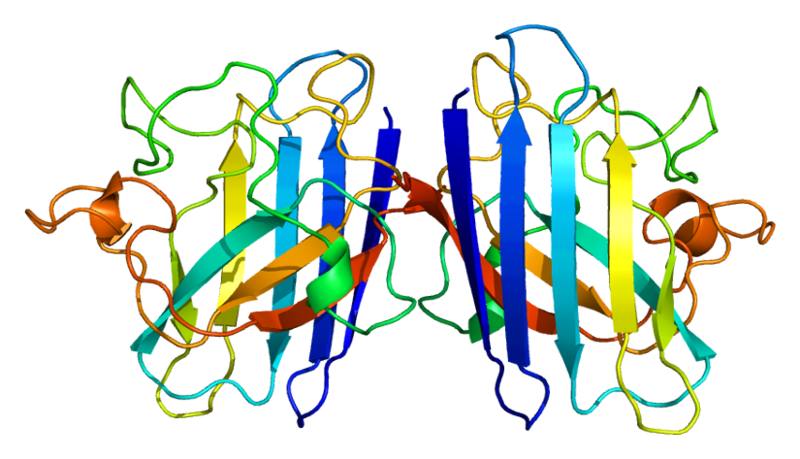
| - | === Superoxide dismutase 1 | + | === Superoxide dismutase 1 (SOD1) === |
| - | [http:// | + | Human soluble Superoxide dismutase 1 (SOD1) is a soluble cytoplasmic protein functional as a homodimer that binds copper and zink ions. SOD1 catalyzes the reaction O<sup>-</sup><sub>2</sub> + O<sup>-</sup><sub>2</sub> + 2H<sup>+</sup> → H<sub>2</sub>O<sub>2</sub> + O<sub>2</sub>, protecting the cell from oxidative damage. SOD1 was first cloned and expressed in ''Escherichia coli'' by [http://www.ncbi.nlm.nih.gov/pubmed/3889846 Hallewell ''et al''., (1985)]. |
| - | + | {|border="1" align="center" cellpadding="3" cellspacing="0" | |
| - | + | !colspan="2"|Gene (cDNA) | |
| - | + | |rowspan="10" width="250"|[[Image:SOD1_dimeric.png|250px]]<br />3D structure of human SOD1 in its dimeric form. Primary citation [http://www.ncbi.nlm.nih.gov/pubmed/20822138 Leinartaite ''et al''. (2010)] | |
| - | [[Image:SOD1_dimeric.png| | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |width="130"|'''Length''' |
| - | | 465 bp | + | |width="130"|465 bp |
|- | |- | ||
| - | | | + | |'''GenBank''' |
| - | | | + | |[http://www.ncbi.nlm.nih.gov/nucleotide/38489879?report=genbank&log$=nucltop&blast_rank=22&RID=CAM83NYN01S AY450286.1]<br /> |
|- | |- | ||
| - | | | + | !colspan="2"|Protein |
| - | + | ||
|- | |- | ||
| - | | ''' | + | |'''Length''' |
| - | | | + | |154 aa |
|- | |- | ||
| - | | | + | |'''Size''' |
| - | | | + | |15,936 Da |
|- | |- | ||
| - | | | + | |'''Fasta''' |
| - | | | + | |[http://www.ncbi.nlm.nih.gov/protein/49456443?report=fasta SOD1]<br /> |
|- | |- | ||
| - | | | + | |colspan="2"|First reported by [http://www.ncbi.nlm.nih.gov/pubmed/3889846 Hallewell ''et al''., (1985)]. |
| - | | [http://www.ncbi.nlm.nih.gov/ | + | |
|} | |} | ||
| - | + | ---- | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
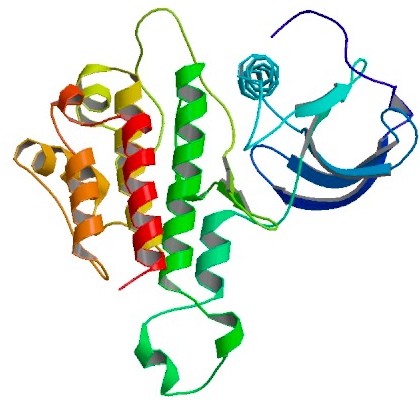
| - | Yeast copper chaperon protein (yCCS) is a | + | === Yeast copper chaperon (yCCS) === |
| + | Yeast copper chaperon protein (yCCS) is a helper chaperon specific for copper/zinc superoxide dismutase located to the cytoplasm. yCCS generates fully metallized, active SOD1 proteins that in turn protects the cell from oxidative damage. | ||
yCCS has been shown to successfully mediate the delivery of copper ions to human SOD1 ([http://www.ncbi.nlm.nih.gov/pubmed/15358352 Ahl ''et al''. 2003]). Co-expression of SOD1 and yCCS yields proteins with higher copper contents, leading to increased activity and more stable proteins. | yCCS has been shown to successfully mediate the delivery of copper ions to human SOD1 ([http://www.ncbi.nlm.nih.gov/pubmed/15358352 Ahl ''et al''. 2003]). Co-expression of SOD1 and yCCS yields proteins with higher copper contents, leading to increased activity and more stable proteins. | ||
| - | |||
| - | |||
| - | + | {|border="1" align="center" cellpadding="3" cellspacing="0" | |
| - | + | !colspan="2"|Gene (cDNA) | |
| - | {| | + | |rowspan="10" width="250"|[[Image:YSOD+yCCS_interaction.jpg|250px]]<br />3D structure of yCCS interacting with yeast superoxide dismutase (ySOD) in it's monomeric form. Ions indicated as gray orbs. Primary citation [http://www.ncbi.nlm.nih.gov/pubmed/11524675 Lamb ''et al''. 2001] |
| - | | | + | |
| - | | | + | |
|- | |- | ||
| - | | | + | |width="130"|'''Length''' |
| - | | | + | |width="130"|750 bp |
|- | |- | ||
| - | | | + | |'''GenBank''' |
| - | | | + | |[http://www.ncbi.nlm.nih.gov/nuccore/NM_001182535.1?report=genbank&log$=seqview NM_001182535.1]<br /> |
|- | |- | ||
| - | | | + | !colspan="2"|Protein |
| - | + | ||
|- | |- | ||
| - | | ''' | + | |'''Length''' |
| - | | | + | |249 aa |
|- | |- | ||
| - | | | + | |'''Size''' |
| - | | | + | |27,330 Da |
|- | |- | ||
| - | | | + | |'''Fasta''' |
| - | | | + | |[http://www.ncbi.nlm.nih.gov/protein/596088?report=fasta yCCS]<br /> |
|- | |- | ||
| - | | | + | |colspan="2"|First reported by [http://www.ncbi.nlm.nih.gov/pubmed/9295278 Culotta ''et al''. (1997)].<br /><br><br><br><br><br> |
| - | | [http://www.ncbi.nlm.nih.gov/ | + | |
|} | |} | ||
| - | + | ---- | |
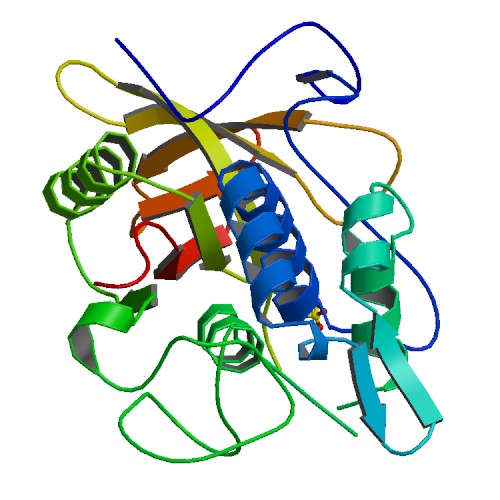
| - | + | === Human basic fibroblast growth factor (bFGF) === | |
| + | {|border="1" align="center" cellpadding="3" cellspacing="0" | ||
| + | !colspan="2"|Gene (cDNA) | ||
| + | |rowspan="10" width="250"|[[Image:BFGF.jpg|250px]]<br />3D structure of bFGF. Primary citation [http://www.ncbi.nlm.nih.gov/pubmed/20133753 Bae ''et al''. 2010] | ||
| + | |- | ||
| + | |width="130"|'''Length''' | ||
| + | |width="130"|468 bp | ||
| + | |- | ||
| + | |'''GenBank''' | ||
| + | |(full mRNA) [http://www.ncbi.nlm.nih.gov/nuccore/153285460 153285460]<br /> | ||
| + | |- | ||
| + | !colspan="2"|Protein | ||
| + | |- | ||
| + | |'''Length''' | ||
| + | |155 aa | ||
| + | |- | ||
| + | |'''Size''' | ||
| + | |17,353 Da | ||
| + | |- | ||
| + | |'''Fasta'''<br /><br><br><br><br> | ||
| + | |[http://www.ncbi.nlm.nih.gov/protein/153285461?report=fasta bFGF]<br /><br><br><br><br> | ||
| + | |} | ||
| + | ---- | ||
| + | === Protein A, z domain === | ||
| + | {|border="1" align="center" cellpadding="3" cellspacing="0" | ||
| + | !colspan="2"|Genepart | ||
| + | |rowspan="10" width="250"|[[Image:ProteinA_z_domain.jpg|250px]]<br />3D structure of the Z-domain of Protein A. Primary citation [http://www.ncbi.nlm.nih.gov/pubmed/9325113 Tashiro ''et al''. 2010] | ||
| + | |- | ||
| + | |width="130"|'''Length''' | ||
| + | |width="130"|174 bp | ||
| + | |- | ||
| + | |'''GenBank''' | ||
| + | |[http://www.ncbi.nlm.nih.gov/gene/2859152 2859152] (full protein)<br /> | ||
| + | |- | ||
| + | !colspan="2"|Protein | ||
| + | |- | ||
| + | |'''Length''' | ||
| + | |58 aa | ||
| + | (508 aa, full protein ) | ||
| + | |- | ||
| + | |'''Size''' | ||
| + | |55,439 Da (full protein) | ||
| + | |- | ||
| + | |'''Fasta'''<br /><br><br><br><br><br> | ||
| + | |[http://www.uniprot.org/uniprot/P38507.fasta Protein A] (full protein)<br /><br><br><br><br><br> | ||
| + | |} | ||
---- | ---- | ||
| + | === IgG protease (IdeS) === | ||
| - | === | + | {|border="1" align="center" cellpadding="3" cellspacing="0" |
| - | + | !colspan="2"|Gene (cDNA) | |
| - | + | |rowspan="10" width="250"|[[Image:IdeS.jpg|250px]]<br />Primary citation [http://www.ncbi.nlm.nih.gov/pubmed/15574492 Wenig ''et al''. 2004] | |
| - | + | ||
| - | + | ||
| - | [[Image: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |width="130"|'''Length''' |
| - | | | + | |width="130"|930 bp |
|- | |- | ||
| - | | | + | |'''GenBank''' |
| - | | | + | |[http://www.ncbi.nlm.nih.gov/gene/6985687 6985687]<br /> |
|- | |- | ||
| - | | | + | !colspan="2"|Protein |
| - | + | ||
|- | |- | ||
| - | | ''' | + | |'''Length''' |
| - | | | + | |339 aa |
|- | |- | ||
| - | | | + | |'''Size''' |
| - | | | + | |37,977 Da |
|- | |- | ||
| - | | | + | |'''Fasta'''<br /><br><br><br><br><br> |
| - | + | |[http://www.ncbi.nlm.nih.gov/protein/209559219?report=fasta IdeS]<br /><br><br><br><br><br> | |
| - | + | ||
| - | + | ||
| - | | [http://www.ncbi.nlm.nih.gov/protein/ | + | |
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 149: | Line 151: | ||
---- | ---- | ||
| - | === | + | == Cell penetrating peptides == |
| - | + | ||
| + | These cell-penetrating peptides, (CPPs) may be used in N- and C-terminal fusions with full-length proteins to create transduction proteins with the ability to permeate the lipid bilayer of various cell types, making them potential gene or protein delivery vectors. | ||
| - | |||
| - | |||
| + | === TAT cell penetrating peptide (TAT) === | ||
| + | Purified full-length TAT fusion proteins expressed in ''Escherichia coli'' have been shown to successfully translocate into several human cell types, including all cells found in whole blood, as well as bone marrow stem cells and osteoblasts, while still retaining the fused protein's activity ([http://www.ncbi.nlm.nih.gov/pubmed/9846587 Nagahara ''et al.'' 1998]). The mechanism for transduction over the bilipid membrane is still a matter of debate, but has been suggested to occur through macropinocytosis, a specialized form of endocytosis ([http://www.ncbi.nlm.nih.gov/pubmed/17913584 Gump and Dowdy, 2007]). | ||
| + | TAT is an 11-amino acid derivative from the Human Immunodeficiency Virus 1 (HIV-1) ''trans''-activating transcriptional activator (Tat) ([http://www.ncbi.nlm.nih.gov/pubmed/2849509 Green and Loewenstein, 1988]; [http://www.ncbi.nlm.nih.gov/pubmed/9846587 Nagahara ''et al.'' 1998]). This part was back translated from the corresponding amino acid sequence and optimized for expression in ''Escherichia coli''. Codon usage has been varied for repetitive amino acids to enable DNA synthesis. | ||
| - | + | {|border="1" align="center" cellpadding="3" cellspacing="0" | |
| - | {| | + | !colspan="2"|Sequence |
| - | | | + | |
| - | + | ||
|- | |- | ||
| - | | | + | |width="130"|'''Length''' |
| - | | | + | |width="130"|33 bp |
|- | |- | ||
| - | | | + | |'''Patent PCT''' |
| - | | | + | |[http://v3.espacenet.com/publicationDetails/biblio;jsessionid=646EDA06997EDDFC0CC04CCE49F87F6B.espacenet_levelx_prod_5?CC=WO&NR=2005084158A2&KC=A2&FT=D&date=20050915&DB=EPODOC&locale=se_se WO 2005/084158 A2]<br /> |
|- | |- | ||
| - | | | + | !colspan="2"|Peptide |
| - | + | ||
|- | |- | ||
| - | | ''' | + | |'''Length''' |
| - | | | + | |11 aa |
|- | |- | ||
| - | | | + | |'''Size''' |
| - | | | + | |1,560 Da ([http://www.scripps.edu/~cdputnam/protcalc.html calculated]) |
|- | |- | ||
| - | | | + | |colspan="2" align="center"|YGRKKRRQRRR |
| - | | | + | |
|- | |- | ||
| - | | | + | |colspan="2"|First reported by |
| - | | | + | [http://www.ncbi.nlm.nih.gov/pubmed/2849509 Green ''et al''. (1988)] and [http://www.ncbi.nlm.nih.gov/pubmed/2849510 Frankel ''et al''. (1988)]<br /> |
|} | |} | ||
| - | + | ---- | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | === Low molecular weight protamine (LMWP) === | ||
| + | Enzymatically prepared LMWP chemically conjugated to ovalbumin (OVA) and bovine serum albumin (BSA) have previously been shown to penetrate the lipid bilayer of human keratinocytes, as well as to successfully permeate mouse skin epidermis ([http://www.ncbi.nlm.nih.gov/pubmed/20232417 Huang ''et al.'', 2010]). Furthermore, LMWP/pDNA complexes can efficiently penetrate into human embryonic kidney cells ([http://www.ncbi.nlm.nih.gov/pubmed/12898639 Park ''et al.'', 2003]). As LMWP has been shown to be neither toxic nor immunogenic ([http://www.ncbi.nlm.nih.gov/pubmed/11741268 Chang ''et al.'' a, 2001]; [http://www.ncbi.nlm.nih.gov/pubmed/11741269 Chang ''et al.'' b, 2001]; [http://www.ncbi.nlm.nih.gov/pubmed/11741270 Lee ''et al.'', 2001]), it may be used as a potential vaccine, drug or gene delivery vector. | ||
| + | LMWP is a 14-amino acid derivative from Rainbow trout (''Oncorhynchus mykiss'') protamine, an arginine-rich protein that replaces histones in chromatin during spermatogenesis ([http://www.ncbi.nlm.nih.gov/pubmed/3755398 McKay ''et al.'', 1986]; [http://www.ncbi.nlm.nih.gov/pubmed/10213181 Byun ''et al.'', 1999]). This part was back translated from the corresponding amino acid sequence and optimized for expression in ''Escherichia coli''. Codon usage has been varied for repetitive amino acids to enable DNA synthesis. | ||
| + | {|border="1" align="center" cellpadding="3" cellspacing="0" | ||
| + | !colspan="2"|Sequence | ||
| + | |- | ||
| + | |width="130"|'''Length''' | ||
| + | |width="130"|42 bp | ||
| + | |- | ||
| + | |'''Patent Application''' | ||
| + | |[http://www.freepatentsonline.com/y2007/0071677.html 20070071677]<br /> | ||
| + | |- | ||
| + | !colspan="2"|Peptide | ||
| + | |- | ||
| + | |'''Length''' | ||
| + | |14 aa | ||
| + | |- | ||
| + | |'''Size''' | ||
| + | |1,880 Da ([http://www.scripps.edu/~cdputnam/protcalc.html calculated]) | ||
| + | |- | ||
| + | |colspan="2" align="center"|VSRRRRRRGGRRRR | ||
| + | |- | ||
| + | |colspan="2"|Patent application by Park et al. (2004)<br /> | ||
| + | |} | ||
---- | ---- | ||
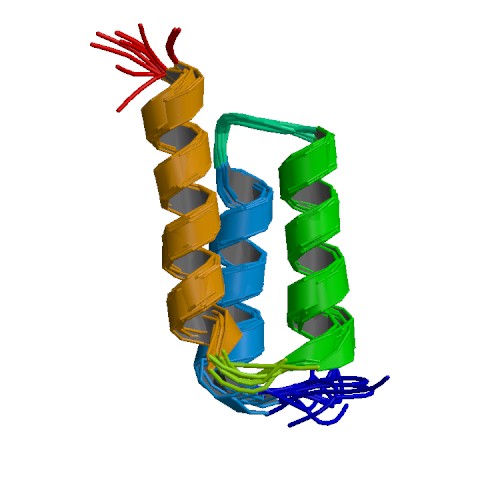
| + | === Transportan 10 (Tp10) === | ||
| + | Chemically synthesized Tp10 peptides conjugated to different cargo, including pDNA and protein, have been shown to efficiently penetrate the lipid bilayer of both human and mouse cells ([http://www.ncbi.nlm.nih.gov/pubmed/15763630 Kilk ''et al.'', 2005]). Membrane permeation is both energy and temperature independent ([http://www.ncbi.nlm.nih.gov/pubmed/11718666 Hällbrink ''et al.'', 2001]). The exact mechanism for penetration is still unclear ([http://www.ncbi.nlm.nih.gov/pubmed/17218466 Yandek ''et al.'', 2007]). | ||
| + | Tp10 is a 21-amino acid derivative from the parent peptide transportan (originally known as galparan), which is a peptide chimera of the neuropeptide galanin and the wasp venom peptide mastoparan ([http://www.ncbi.nlm.nih.gov/pubmed/10930519 Soomets ''et al.'', 2000]; [http://www.ncbi.nlm.nih.gov/pubmed/8738882 Langel ''et al.'', 1996]). This part was back translated from the corresponding amino acid sequence and optimized for expression in ''Escherichia coli''. Codon usage has been varied for repetitive amino acids to enable DNA synthesis. | ||
| - | === | + | {|border="1" align="center" cellpadding="3" cellspacing="0" |
| - | + | !colspan="2"|Sequence | |
| - | + | |rowspan="8" width="250"|[[Image:Transportan.jpg|250px]]<br />3D structure of transportan<br /> [http://www.dbb.su.se/Faculty/Lena_M%C3%A4ler/Structural_basis_for_peptide-membrane_interactions www.dbb.su.se] | |
| - | + | ||
| - | + | ||
| - | [[Image: | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |width="130"|'''Length''' |
| - | | | + | |width="130"|63 bp |
|- | |- | ||
| - | | | + | |'''Patent Application''' |
| - | | | + | |[http://www.freepatentsonline.com/y2008/0234183.html 20080234183]<br /> |
|- | |- | ||
| - | | | + | !colspan="2"|Peptide |
| - | + | ||
|- | |- | ||
| - | | ''' | + | |'''Length''' |
| - | | | + | |21 aa |
|- | |- | ||
| - | | | + | |'''Size''' |
| - | | | + | |2,183 Da ([http://www.scripps.edu/~cdputnam/protcalc.html calculated]) |
|- | |- | ||
| - | | | + | |colspan="2" align="center"|AGYLLGKINLKALAALAKKIL |
| - | | | + | |
|- | |- | ||
| - | | | + | |colspan="2"|Patent application by Hallbrink et al. (2003)<br /> |
| - | | | + | |
|} | |} | ||
| - | + | <!--|- | |
| - | + | |rowspan="8" width="250"|[[Image:Tp10_prediction.png|250px]]<br />3D structure of transportan<br /> [http://www.dbb.su.se/Faculty/Lena_M%C3%A4ler/Structural_basis_for_peptide-membrane_interactions www.dbb.su.se]--> | |
| - | + | ---- | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
{{Stockholm/Footer}} | {{Stockholm/Footer}} | ||
Latest revision as of 01:19, 28 October 2010
 |

|
 |

|
 |

|

|

|
 "
"