Team:Michigan/Pili August September
From 2010.igem.org
(→8/23/2010) |
(→8/18/2010) |
||
| (3 intermediate revisions not shown) | |||
| Line 120: | Line 120: | ||
'''Miniprep pBAD plasmid''' | '''Miniprep pBAD plasmid''' | ||
| - | # | + | #Inoculate 5.0mL LB in 50mL conical tubes w/ 100ug/mL of ampilicin |
| - | ## | + | ##The cultures (2 of them) grew for ~12hrs (8am to 8pm). |
| - | # | + | #Centrifuge (using the 50mL conical tubes) for 5000rpm for 10min at 4ºC |
| - | # | + | #Carefully discard the supernatant and resuspend the pellet with 250uL of P1 buffer (kept in the 4ºC fridge) |
| - | # | + | #Transfer into a labelled 1.5mL eppendorf tube (set pipetman to 500uL just in case) |
| - | # | + | #Add 250uL P2 Buffer--> invert 4-6times (should turn blue)--> add 350uL N3 buffer--> invert 4-6times (should become vicious or clumpy) |
| - | ## | + | ##Centrifuge at 13,000 rpm for 10 min |
| - | # | + | #Pipette out supernatant into a QIA spin column--> centrifuge for 60s--> discard flow through |
##did not pipet out all of the supernatant | ##did not pipet out all of the supernatant | ||
| - | # | + | #Add 500uL PB buffer --> centrifuge 60s--> discard flow through |
| - | # | + | #Add 750uL PE buffer--> centrifuge 60s--> discard--> centrifuge 60s again |
| - | # | + | #Transfer into a labeled 1.5mL eppendorf tube--> add 50uL EB (elution buffer) |
| - | ## | + | ##Allow it to sit for 1 min (it helps to release the DNA from the column) |
'''fimB PCR product Purification''' | '''fimB PCR product Purification''' | ||
#Used sample A and B of PCR product (save C and D for later) | #Used sample A and B of PCR product (save C and D for later) | ||
| - | ## | + | ##Total volume of PCR product = 81 uL (40.5uL separately) |
| - | # | + | #Add 5 volumes of PB buffer (202.5uL) to 1 volume of PCR product (40.5uL); invert |
| - | # | + | #Transfer into a QIAquick spin column (provided in the Qiagen kit) |
| - | ## | + | ##Set pipetman to 260uL to be sure to get all of the mixture |
| - | # | + | #Centrifuge spin column at 13,000rpm for 60s |
| - | # | + | #Discard the flow through (in the collection tube) and add 750uL PE buffer (to wash the DNA) |
| - | + | Repeat step 4 again | |
| - | # | + | #Discard flow through and centrifuge again to get the remain buffers out |
| - | # | + | #Place the column into a labeled 1.5mL eppendorf tube |
| - | # | + | #Add 50uL EB buffer (to elute out the DNA) directly to the white inner circle of the column (avoid touching the pipet tip to the column) |
| - | ## | + | ##Allow the mix to sit in the column for 1 min, then centrifuge for 1 min (13,000rpm) |
| - | ## | + | ##Remember to point the cap of the eppendorf tube in the opposite direction of the centrifuge machine |
'''fimB Digest''' | '''fimB Digest''' | ||
| - | # | + | #Pre-programmed PCR machine to Digest (DIG1) |
| - | # | + | #Use 5 PCR reaction tubes for each; 5 for fimB and 5 for pBAD |
| - | # | + | #Add the following amounts in that order (total volume of 20uL) |
*16 uL of the DNA (fimB and pBAD to their respective tubes) | *16 uL of the DNA (fimB and pBAD to their respective tubes) | ||
*2 uL of NEB 2 buffer | *2 uL of NEB 2 buffer | ||
*1 uL NcoI | *1 uL NcoI | ||
*1uL HindIII | *1uL HindIII | ||
| - | # | + | #Incubate for 37ºC overnight (12hrs) |
| - | ## | + | ##Place into 4ºC fridge the next day |
==8/16/2010== | ==8/16/2010== | ||
''Kevin, Marc, Alena'' | ''Kevin, Marc, Alena'' | ||
| - | Alena order/picks up NEB Ligase (T4 DNA Ligase, 20,000U/mL) from MSRB II enzyme store | + | Alena order/picks up NEB Ligase (T4 DNA Ligase, 20,000U/mL) from MSRB II enzyme store. |
Attempted experiment: | Attempted experiment: | ||
*Added 1uL of CIP (Calf Intestine Phosphatase) to plasmid reactions only (pBAD) | *Added 1uL of CIP (Calf Intestine Phosphatase) to plasmid reactions only (pBAD) | ||
**CIP will cut off the 3' phosphate to prevent the plasmid from folding back on itself | **CIP will cut off the 3' phosphate to prevent the plasmid from folding back on itself | ||
| - | * | + | *Incubate CIP-pBAD at 37C for 1 hr and then heat-shock (65ºC for 15 min) |
'''We made a huge human error when setting up our thermal cycle program on the PCR machine. We did not realize we only set the thermal cycle for our digest on 8/14/2010>> [https://2010.igem.org/Team:Michigan/Pili_Expression#8.2F14.2F2010] for 12 minutes (12:00) rather than the intended time period of 12 hours (12:00:00). This explains the condensation present in the PCR machine (it stayed at 4C for too long). We discovered our mistake after running the incubation of CIP+pBAD for 37C for (1:00) 1 minute then heat-shock at 65C for 15 minute. It is VERY IMPORTANT to triple check one another when entering in any program.''' | '''We made a huge human error when setting up our thermal cycle program on the PCR machine. We did not realize we only set the thermal cycle for our digest on 8/14/2010>> [https://2010.igem.org/Team:Michigan/Pili_Expression#8.2F14.2F2010] for 12 minutes (12:00) rather than the intended time period of 12 hours (12:00:00). This explains the condensation present in the PCR machine (it stayed at 4C for too long). We discovered our mistake after running the incubation of CIP+pBAD for 37C for (1:00) 1 minute then heat-shock at 65C for 15 minute. It is VERY IMPORTANT to triple check one another when entering in any program.''' | ||
Rest of the day: | Rest of the day: | ||
| - | * | + | *Headed over to the ERB and made two cultures of 5mL LB-Amp (in 50mL conical tubes)--> placed in the Lin Lab 4ºC fridge |
==8/17/2010== | ==8/17/2010== | ||
| Line 207: | Line 207: | ||
Met with Chris to discuss about yeast agglutination | Met with Chris to discuss about yeast agglutination | ||
| - | *Chris sent us a possible paper with a decent protocol to look at for the assay [[Media:YeastAgglutinationPaper.pdf]] | + | *Chris sent us a possible paper with a decent protocol to look at for the assay [[Media:YeastAgglutinationPaper.pdf]]. |
'''NOTE ABOUT K.O. STRAINS''' | '''NOTE ABOUT K.O. STRAINS''' | ||
| Line 339: | Line 339: | ||
''Kevin'' | ''Kevin'' | ||
| - | Created frozen stocks of K12 and ΔFim::kan w/the plasmid from overnight cultures. Stored them in box 1 in the ERB - | + | Created frozen stocks of K12 and ΔFim::kan w/the plasmid from overnight cultures. Stored them in box 1 in the ERB -20ºC freezer. |
Ran miniprep of K12 and ΔFim::kan, using 5 ml of each culture. | Ran miniprep of K12 and ΔFim::kan, using 5 ml of each culture. | ||
Note: Centrifuge in ERB will only go up to 5000 rpm w/50 ml tubes, therefore we transferred the culture to several eppendorf tubes. This was probably not a good idea, because we were left with a lot of leftover supernatant and that could have diluted the buffers. In the future, we should centrifuge the culture 1 ml at a time, and add 1 ml after each cycle. | Note: Centrifuge in ERB will only go up to 5000 rpm w/50 ml tubes, therefore we transferred the culture to several eppendorf tubes. This was probably not a good idea, because we were left with a lot of leftover supernatant and that could have diluted the buffers. In the future, we should centrifuge the culture 1 ml at a time, and add 1 ml after each cycle. | ||
| - | Stored the product from miniprep in box 2 in the ERB - | + | Stored the product from miniprep in box 2 in the ERB -20ºC freezer. |
==9/9/2010== | ==9/9/2010== | ||
Latest revision as of 03:05, 27 October 2010
| Sunday | Monday | Tuesday | Wednesday | Thursday | Friday | Saturday | |
| Week 6 | - | - | - | - | - | - | 8/7/2010 |
| Week 7 | - | 8/9/2010 | - | 8/11/2010 | 8/12/2010 | - | 8/14/2010 |
| Week 8 | - | 8/16/2010 | 8/17/2010 | 8/18/2010 | - | - | - |
| Week 9 | 8/22/2010 | 8/23/2010 | - | - | - | - | - |
| Week 10 | - | - | - | - | 9/2/2010 | - | - |
| Week 11 | - | - | 9/7/2010 | 9/8/2010 | 9/9/2010 | 9/10/2010 | - |
8/7/2010
Kevin, Marc, Alena
PCR #1 Used a gradient from 40ºC to 60ºC for the first 3 cycles to find the optimum annealing temperature.
All of the annealing temperatures gave a good result according to the gel.
8/9/2010
Kevin, Marc, Alena
Used a 57ºC degree annealing temperature to get enough DNA for the digest and ligation.
4 out of the 5 PCR reactions worked well according to the gel.
The 5th well could have been a loading problem or there wasn't enough DNA.
8/11/2010
Kevin, Marc
Met with Chris, received advice for updating digest and ligation protocols.
8/12/2010
Kevin, Marc, Alena
Met and discussed protocols for digestion and ligation of FIMB into pBAD.
1. Added 5 mL of LB broth each to 2 50 mL falcon tubes in the ERB lab using sterile technique.
2. Added 5 microliters of Kanamycin to each of the 50 mL tubes in step 1.
Went to the budget committee meeting for 1 hour with the tubes.
3. Obtained the cryostock of pBAD from the Lin -80C freezer (iGEM box cell #73)
4. Stabbed cryostock using a sterile 200 microliter pipette tip and pipetted into media from step 2.
5. At 8:05PM placed the two falcon tubes from step 4 into the incubator/shaker at 30ºC.
8/14/2010
Kevin, Marc, Alena
Miniprep pBAD plasmid
- Inoculate 5.0mL LB in 50mL conical tubes w/ 100ug/mL of ampilicin
- The cultures (2 of them) grew for ~12hrs (8am to 8pm).
- Centrifuge (using the 50mL conical tubes) for 5000rpm for 10min at 4ºC
- Carefully discard the supernatant and resuspend the pellet with 250uL of P1 buffer (kept in the 4ºC fridge)
- Transfer into a labelled 1.5mL eppendorf tube (set pipetman to 500uL just in case)
- Add 250uL P2 Buffer--> invert 4-6times (should turn blue)--> add 350uL N3 buffer--> invert 4-6times (should become vicious or clumpy)
- Centrifuge at 13,000 rpm for 10 min
- Pipette out supernatant into a QIA spin column--> centrifuge for 60s--> discard flow through
- did not pipet out all of the supernatant
- Add 500uL PB buffer --> centrifuge 60s--> discard flow through
- Add 750uL PE buffer--> centrifuge 60s--> discard--> centrifuge 60s again
- Transfer into a labeled 1.5mL eppendorf tube--> add 50uL EB (elution buffer)
- Allow it to sit for 1 min (it helps to release the DNA from the column)
fimB PCR product Purification
- Used sample A and B of PCR product (save C and D for later)
- Total volume of PCR product = 81 uL (40.5uL separately)
- Add 5 volumes of PB buffer (202.5uL) to 1 volume of PCR product (40.5uL); invert
- Transfer into a QIAquick spin column (provided in the Qiagen kit)
- Set pipetman to 260uL to be sure to get all of the mixture
- Centrifuge spin column at 13,000rpm for 60s
- Discard the flow through (in the collection tube) and add 750uL PE buffer (to wash the DNA)
Repeat step 4 again
- Discard flow through and centrifuge again to get the remain buffers out
- Place the column into a labeled 1.5mL eppendorf tube
- Add 50uL EB buffer (to elute out the DNA) directly to the white inner circle of the column (avoid touching the pipet tip to the column)
- Allow the mix to sit in the column for 1 min, then centrifuge for 1 min (13,000rpm)
- Remember to point the cap of the eppendorf tube in the opposite direction of the centrifuge machine
fimB Digest
- Pre-programmed PCR machine to Digest (DIG1)
- Use 5 PCR reaction tubes for each; 5 for fimB and 5 for pBAD
- Add the following amounts in that order (total volume of 20uL)
- 16 uL of the DNA (fimB and pBAD to their respective tubes)
- 2 uL of NEB 2 buffer
- 1 uL NcoI
- 1uL HindIII
- Incubate for 37ºC overnight (12hrs)
- Place into 4ºC fridge the next day
8/16/2010
Kevin, Marc, Alena
Alena order/picks up NEB Ligase (T4 DNA Ligase, 20,000U/mL) from MSRB II enzyme store.
Attempted experiment:
- Added 1uL of CIP (Calf Intestine Phosphatase) to plasmid reactions only (pBAD)
- CIP will cut off the 3' phosphate to prevent the plasmid from folding back on itself
- Incubate CIP-pBAD at 37C for 1 hr and then heat-shock (65ºC for 15 min)
We made a huge human error when setting up our thermal cycle program on the PCR machine. We did not realize we only set the thermal cycle for our digest on 8/14/2010>> [1] for 12 minutes (12:00) rather than the intended time period of 12 hours (12:00:00). This explains the condensation present in the PCR machine (it stayed at 4C for too long). We discovered our mistake after running the incubation of CIP+pBAD for 37C for (1:00) 1 minute then heat-shock at 65C for 15 minute. It is VERY IMPORTANT to triple check one another when entering in any program.
Rest of the day:
- Headed over to the ERB and made two cultures of 5mL LB-Amp (in 50mL conical tubes)--> placed in the Lin Lab 4ºC fridge
8/17/2010
Kevin, Marc, Alena
Marc inoculated the two cultures made last night (8/16/2010) with pBAD and grew in 37C shaker (~9am to ~6p, 9hrs)
NOTE REGARDING LB CULTURE MADE ON 8/16/2010
- Marc noticed the 400mL LB media made in the ERB to be contaminated.
- Our two cultures could have been contaminated but pBAD grew pretty well (saturated) so we're not too worried
fimE and fimB knock out (K.O.) strains came in
- Marc adds 50uL of kanamycin onto each LB-agar plate and spread evenly with sterile glass beads
- LB-agar plate ~25mL
- Jeremy Minty's advice: more Kan is better than too little --> Kan 100 instead of Kan 50
- allow the plate to absorb the kanamycin for 2 hours before applying the strains
- Alena follows Jeremy Minty's example of carefully taking out the small filter circle out of the foil with sterile techniques onto a LB-agar plate (previously labeled)
- add ~75uL of LB onto the filter circle
- streak the filter paper w/ inoculating streakers (used 3 of them)
- incubate plates w/ filter paper (still on the agar) upside (agar side up) at 37C overnight
Followed 8/14/2010 protocol (above) for:
- Miniprep for pBAD plasmid
- Last time we used the incorrect spin column (QIAquick spin column; purple; used for PCR purification). This time we used QIAprep spin column (blue; had no cap on the column)
- PCR purification for fimB
- used PCR products labeled C and D tubes
- Digest fimB and pBAD
- This time, made sure the time on the program was set to 12:00:00
- 5 rxns for each fimB and pBAD (10 total tubes, 20uL total volume in each tube)
8/18/2010
Kevin, Marc, Alena
Met with Chris to discuss about yeast agglutination
- Chris sent us a possible paper with a decent protocol to look at for the assay Media:YeastAgglutinationPaper.pdf.
NOTE ABOUT K.O. STRAINS
- fimE K.O. did not grow out (which contradicts the prediction that fimE K.O grows faster than fimB K.O.)
- try letting the plate grow at 37ºC for a longer period of time (one colony observed)
- remove the filter circle paper and put onto a new LB-agar plate
- suspect too much kanamycin on the plate
Lab work:
- add CIP (calf intestine phosphatase= cuts off the 3' phosphate to prevent plasmid from closing back on itself) to plasmid (pBAD) reaction tubes only--> incubate at 37ºC for 1 hour (using PCR machine)
- heat/inactivate pBAD and fimB reaction tubes for 15 min at 65C (again using PCR machine)
- perform DNA purification on the digests (follow 8/16/2010 procedure)
- using 210uL and 200uL of PB buffer for pBAD and fimB respectively
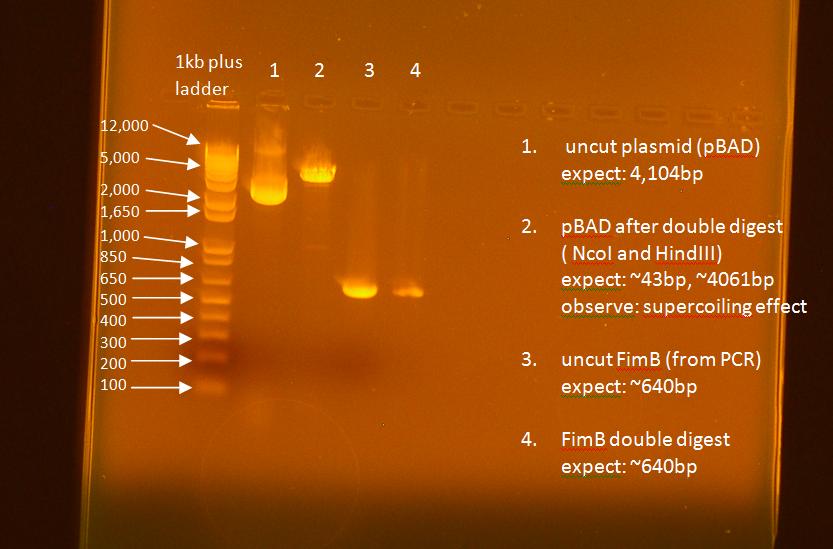
- ran a gel on the digest
- ran nanodrop3.0.1 on the digest
- use EB buffer as the blank
- ALWAYS clean twice when done using the machine
| FimB Digest | pBAD Digest | |
|---|---|---|
| 260/280 | 1.86 | 1.90 |
| 260/230 | 1.83 | 2.20 |
| ng/uL | 24.8 | 59.7 |
8/22/2010
Kevin, Marc
Ran Ligation of FimB and pBAD for 16 hrs O/N
Prepared culture of DH5α for electroporation tomorrow.
8/23/2010
Kevin, Marc
Growing competent cells for electroporation.
Colony started growing at 2:00.
Measured ODs
| Time | OD600 |
|---|---|
| 3:30 | .114 |
| 5:30 | .206 |
| 6:30 | .441 |
Precipitate Ligation product w/butanol
Add to eppendorf tube:
- 50 ul ultrapure H2O
- Ligation product
- 500 ul butanol
Spin at 4ºC, 13000 rpm for 20 min
Made ampicillin plates
- Add 25 ul Amp100 to LB plates
6 plates:
- Cells only
- Plasmid only
- 2x pBAD+FimB undiluted
- 2x pBAD+FimB 1:100
9/2/2010
Kevin
Electroporation of pBAD+FimB into K12 and ΔFimB::kan cells.
15 plates
1x K12 control (amp)
1x Plasmid control (amp + kan)
1x ΔFimB control (amp + kan)
6x ΔFimB (amp + kan)
6x K12 (amp)
| Cell Type | Time Constant |
|---|---|
| Plasmid Control | 5.6 |
| ΔFimB Control | 5.4 |
| K12 Control | 5.6 |
| ΔFimB A | 5.4 |
| ΔFimB B | 5.4 |
| K12 A | 5.8 |
| K12 B | 5.6 |
9/7/2010
Kevin
Started overnight cultures of 1:1000 dilutions of K12 and ΔFim::kan, both with the pBAD+FimB plasmid.
9/8/2010
Kevin
Created frozen stocks of K12 and ΔFim::kan w/the plasmid from overnight cultures. Stored them in box 1 in the ERB -20ºC freezer.
Ran miniprep of K12 and ΔFim::kan, using 5 ml of each culture. Note: Centrifuge in ERB will only go up to 5000 rpm w/50 ml tubes, therefore we transferred the culture to several eppendorf tubes. This was probably not a good idea, because we were left with a lot of leftover supernatant and that could have diluted the buffers. In the future, we should centrifuge the culture 1 ml at a time, and add 1 ml after each cycle.
Stored the product from miniprep in box 2 in the ERB -20ºC freezer.
9/9/2010
Kevin
Cryopreserved frozen stocks of K12 and ΔFimB::kan w/the plasmid.
Ran digest to determine whether the plasmid and FimB were actually in the cell. Used protocol from previous digest on 8/14.
9/10/2010
Kevin
Ran gel of previous day's digest, unfortunately the gel was inconclusive. There is an undetermined error with the gel electrophoresis machine.
 "
"