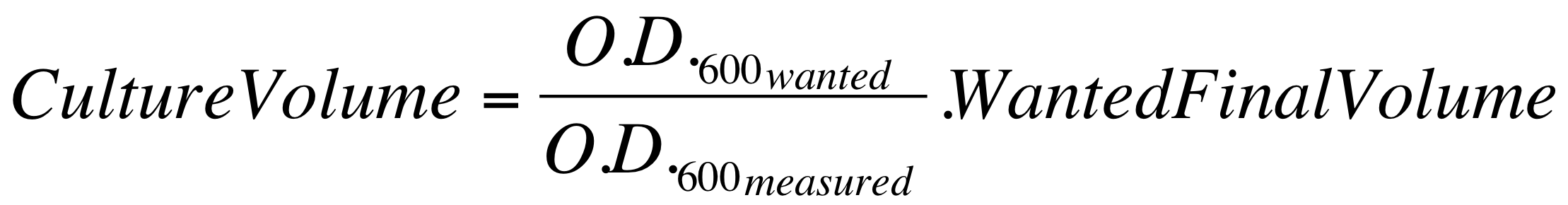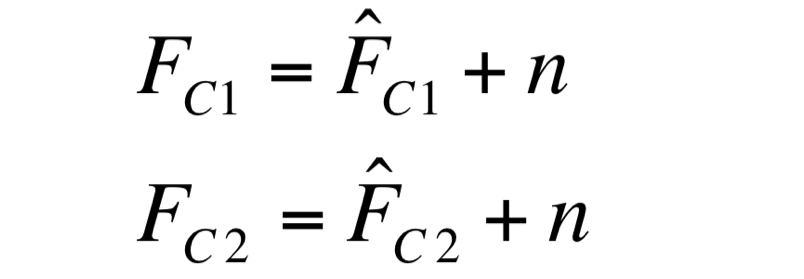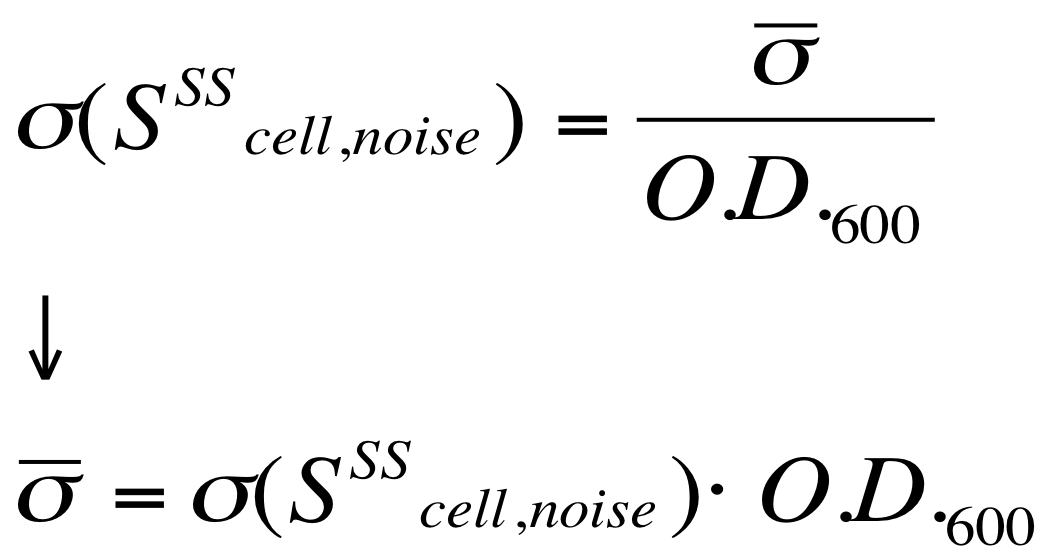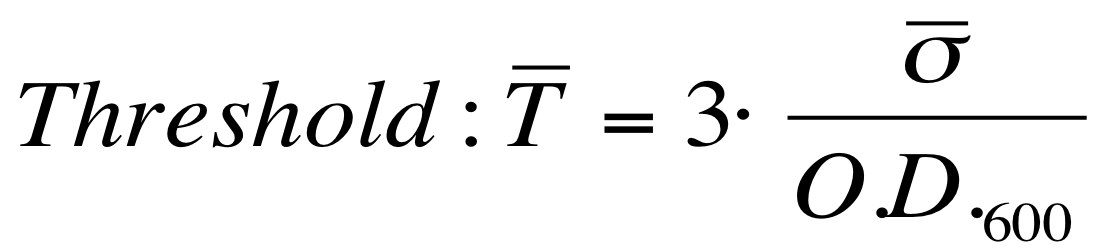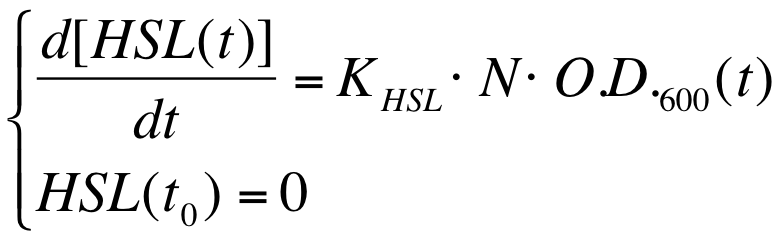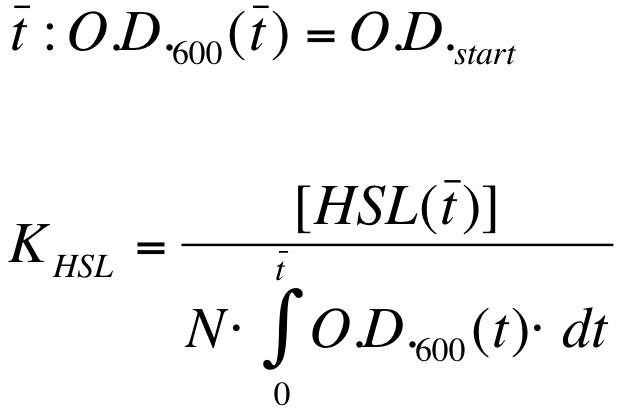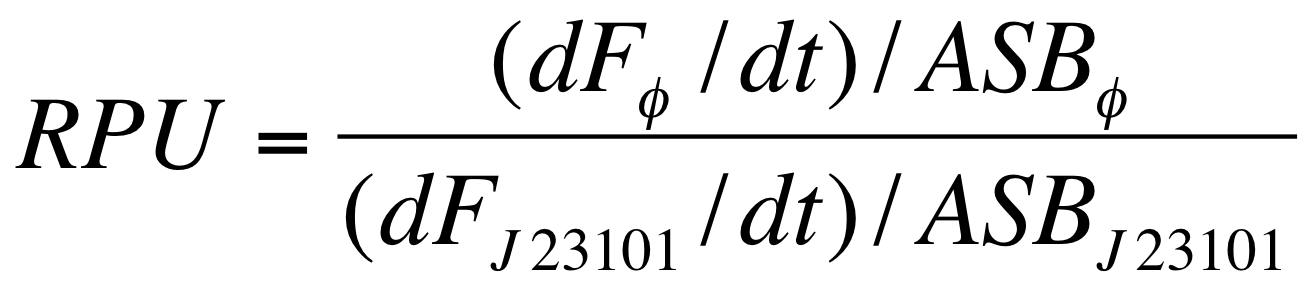Team:UNIPV-Pavia/Parts/Characterization
From 2010.igem.org
(→New Parts) |
|||
| (32 intermediate revisions not shown) | |||
| Line 38: | Line 38: | ||
==[[Team:UNIPV-Pavia/Parts/Characterization/NewParts |New Parts]]== | ==[[Team:UNIPV-Pavia/Parts/Characterization/NewParts |New Parts]]== | ||
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts #BBa_K300000 - BioBrick integrative base vector for E. coli |BBa_K300000 - BioBrick integrative base vector for E. coli]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300000 - BioBrick integrative base vector for E. coli |BBa_K300000 - BioBrick integrative base vector for E. coli]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts #BBa_K300001 - BioBrick integrative base vector for S. cerevisiae|BBa_K300001 - BioBrick integrative base vector for S. cerevisiae | + | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300001 - BioBrick integrative base vector for S. cerevisiae|BBa_K300001 - BioBrick integrative base vector for S. cerevisiae]] |
| - | + | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300010 - PoPS-based self-inducible device|BBa_K300010 - PoPS-based self-inducible device]] | |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts #BBa_K300010 - PoPS-based self-inducible device|BBa_K300010 - PoPS-based self-inducible device]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts#BBa_K300093, BBa_K300094, BBa_K300097, BBa_K300095, BBa_K300086 and BBa_K300084 - Phasin and Intein-based tags for protein purification|BBa_K300093, BBa_K300094, BBa_K300097, BBa_K300095, BBa_K300086 and BBa_K300084 - Phasin and Intein-based tags for protein purification]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/NewParts #BBa_K300093, BBa_K300094, BBa_K300097, BBa_K300095 and BBa_K300084 - Phasin and Intein-based | + | |
| - | + | ||
==[[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts |Improved Parts]]== | ==[[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts |Improved Parts]]== | ||
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300002 - Phasin (PhaP) - head domain|BBa_K300002 - Phasin (PhaP) - head domain]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300002 - Phasin (PhaP) - head domain|BBa_K300002 - Phasin (PhaP) - head domain - improvement of BBa_K208001]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300003 - Phasin (PhaP) - internal domain|BBa_K300003 - Phasin (PhaP) - internal domain]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/RebExistingParts #BBa_K300003 - Phasin (PhaP) - internal domain|BBa_K300003 - Phasin (PhaP) - internal domain - improvement of BBa_K208001]] |
| - | + | ||
==[[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry |Existing Parts from the Registry]]== | ==[[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry |Existing Parts from the Registry]]== | ||
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry #BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device|BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_R0010, BBa_R0011 - Wild type and hybrid lac promoters|BBa_R0010, BBa_R0011 - Wild type and hybrid lac promoters]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry #BBa_F2620 - 3OC6HSL -> PoPS Receiver|BBa_F2620 - 3OC6HSL -> PoPS Receiver]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device|BBa_K300009/BBa_I4102 - PoPS->3OC6HSL sender device]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry #BBa_J61001 - | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_F2620 - 3OC6HSL -> PoPS Receiver|BBa_F2620 - 3OC6HSL -> PoPS Receiver]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry #BBa_J23100, BBa_J23101, BBa_J23105, BBa_J23106, BBa_J23110, BBa_J23114, BBa_J23116, BBa_J23118 - constitutive Anderson | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J61001 - R6K Origin of replication|BBa_J61001 - R6K Origin of replication]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry #BBa_P1004 - | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J23100, BBa_J23101, BBa_J23105, BBa_J23106, BBa_J23110, BBa_J23114, BBa_J23116, BBa_J23118 - constitutive promoters from Anderson's collection|BBa_J23100, BBa_J23101, BBa_J23105, BBa_J23106, BBa_J23110, BBa_J23114, BBa_J23116, BBa_J23118 - constitutive promoters from Anderson's collection]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry #BBa_K125500 - | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_P1004 - chloramphenicol resistance cassette|BBa_P1004 - chloramphenicol resistance cassette]] |
| - | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry #BBa_J72008 - phi80 integration helper plasmid pInt80-649|BBa_J72008 - phi80 integration helper plasmid pInt80-649]] | + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_K125500 - GFP fusion brick|BBa_K125500 - GFP fusion brick]] |
| + | * [[Team:UNIPV-Pavia/Parts/Characterization/ExistingPartsRegistry#BBa_J72008 - phi80 integration helper plasmid pInt80-649|BBa_J72008 - phi80 integration helper plasmid pInt80-649]] | ||
<hr><br> | <hr><br> | ||
| Line 70: | Line 68: | ||
---- | ---- | ||
<br> | <br> | ||
| - | |||
=Growth conditions= | =Growth conditions= | ||
| Line 77: | Line 74: | ||
* 8 ul of long term storage glycerol stock were inoculated in 1 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | * 8 ul of long term storage glycerol stock were inoculated in 1 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
* The grown cultures were then diluted 1:100 in 1 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | * The grown cultures were then diluted 1:100 in 1 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | ||
| - | * | + | * Cultures were then pelletted (2000 rpm, 10 minutes) in order to eliminate the HSL produced during the growth. |
* Supernatants were discarded and the pellets were resuspended in 1 ml of LB or M9 + suitable antibiotic and transferred to a 1.5 ml tube | * Supernatants were discarded and the pellets were resuspended in 1 ml of LB or M9 + suitable antibiotic and transferred to a 1.5 ml tube | ||
| - | * These cultures were diluted 1:1000 in 1 ml of LB or M9 + suitable antibiotic and aliquoted in a flat-bottom 96-well microplate in triplicate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects | + | * These cultures were diluted 1:1000 in 1 ml of LB or M9 + suitable antibiotic and aliquoted in a flat-bottom 96-well microplate in triplicate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects, see https://2009.igem.org/Team:UNIPV-Pavia/Methods_Materials/Evaporation). All the wells were filled with a 200 ul volume. |
* The microplate was incubated in the Tecan Infinite F200 microplate reader and fluorescence and absorbance were measured with this automatic protocol: | * The microplate was incubated in the Tecan Infinite F200 microplate reader and fluorescence and absorbance were measured with this automatic protocol: | ||
** 37°C constant for all the experiment; | ** 37°C constant for all the experiment; | ||
| Line 98: | Line 95: | ||
*8 ul of long term storage glycerol stock were inoculated in 5 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | *8 ul of long term storage glycerol stock were inoculated in 5 ml of LB or M9 + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
*The grown cultures were then diluted 1:100 in 5 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | *The grown cultures were then diluted 1:100 in 5 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours. | ||
| - | *These new cultures were diluted to an O.D.600 of 0.02 (measured with a TECAN F200 microplate reader on a 200 ul of volume per well; it is not the 1 cm pathlength cuvette) in 2 ml LB or M9 + suitable antibiotic. In order to have the cultures at the desired O.D.600, the following dilution was performed: | + | *These new cultures were diluted to an O.D.600 of 0.02 (measured with a TECAN F200 microplate reader on a 200 ul of volume per well; it is not equivalent to the 1 cm pathlength cuvette) in 2 ml (wanted final volume) LB or M9 + suitable antibiotic. In order to have the cultures at the desired O.D.600 (O.D._wanted=0.02), the following dilution was performed: |
[[Image:UNIPV_Pavia_OD600_dil.png|500px|center]] | [[Image:UNIPV_Pavia_OD600_dil.png|500px|center]] | ||
*These new dilutions were aliquoted in a flat-bottom 96-well microplate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects in the frame). All the wells were filled with a 200 ul volume. | *These new dilutions were aliquoted in a flat-bottom 96-well microplate, avoiding to perform dynamic experiments in the microplate frame (in order to prevent evaporation effects in the frame). All the wells were filled with a 200 ul volume. | ||
| Line 107: | Line 104: | ||
**O.D. filter was 600 nm; | **O.D. filter was 600 nm; | ||
**GFP filters were 485nm (ex) / 540nm (em); | **GFP filters were 485nm (ex) / 540nm (em); | ||
| - | ** | + | **RFP filters were 535nm (ex) / 620nm (em); |
**15 seconds of linear shaking (3mm amplitude) followed by 10 seconds of waiting before the measurements in order to make a homogeneous culture. | **15 seconds of linear shaking (3mm amplitude) followed by 10 seconds of waiting before the measurements in order to make a homogeneous culture. | ||
**Experiment duration time: about 6 hours. | **Experiment duration time: about 6 hours. | ||
| Line 124: | Line 121: | ||
**8 ul of BBa_J231xx-<partinfo>BBa_K300009</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | **8 ul of BBa_J231xx-<partinfo>BBa_K300009</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
**The grown cultures were then diluted 1:100 in 5 ml of LB supplemented medium and incubated in the same conditions as before for 6 hours. | **The grown cultures were then diluted 1:100 in 5 ml of LB supplemented medium and incubated in the same conditions as before for 6 hours. | ||
| - | ** | + | **Each falcon was pelletted (2000 rpm, 10 minutes) and supernatants were collected and filtered (0.2 um), in order to eliminate bacterial residues. These supernatants contained the 3OC6-HSL at the concentration produced by cultures. These supernatants were used to "induce" <partinfo>BBa_T9002</partinfo> cultures, in order to quantify the [HSL]. They were conserved at -20°C until the next day. |
| - | + | ||
*<em>Growth and preparation of the biosensor culture of <partinfo>BBa_T9002</partinfo></em> | *<em>Growth and preparation of the biosensor culture of <partinfo>BBa_T9002</partinfo></em> | ||
**8 ul of <partinfo>BBa_T9002</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | **8 ul of <partinfo>BBa_T9002</partinfo> long term glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours. | ||
| Line 143: | Line 139: | ||
*** 0,1 nM | *** 0,1 nM | ||
*** 0 M | *** 0 M | ||
| - | **All inductions were performed adding 2ul of "inducer solution" (3OC6-HSL, Sigma Aldrich) at | + | **All inductions were performed adding 2ul of "inducer solution" (3OC6-HSL, Sigma Aldrich) at the proper concentration in 200 ul of culture. |
*<em>HSL quantification</em> | *<em>HSL quantification</em> | ||
**2ul of supernatants prepared as described above were used to induce 200 ul of <partinfo>BBa_T9002</partinfo> cultures in triplicate. | **2ul of supernatants prepared as described above were used to induce 200 ul of <partinfo>BBa_T9002</partinfo> cultures in triplicate. | ||
| - | **If the amount of 3OC6-HSL present in the supernatant was not enough concentrated to trigger the induction of <partinfo>BBa_T9002</partinfo>, a bigger amount of supernatant (X ul) was used to induce the cultures (200-X ul of cultures). | + | **If the amount of 3OC6-HSL present in the supernatant was not enough concentrated to trigger the induction of <partinfo>BBa_T9002</partinfo>, a bigger amount of supernatant (X ul) was used to induce the cultures (200-X ul of cultures). Then, the calibration curve was obtained by using the same amount of <partinfo>BBa_T9002</partinfo> culture (200-X ul) induced with X ul of inducer solution at the proper concentration. To maintain the same experimental conditions, the inducer was diluted in the supernatant of a negative control (i.e., <partinfo>BBa_B0034</partinfo>). |
**Fluorescence and absorbance were measured after 30 min from the induction with a Tecan Infinite F200 microplate reader and, using the information provided by the calibration curve, the amount of 3OC6-HSL present in every supernatant was evaluated (each reported fluorescence value was corrected with the O.D.600 of the culture). | **Fluorescence and absorbance were measured after 30 min from the induction with a Tecan Infinite F200 microplate reader and, using the information provided by the calibration curve, the amount of 3OC6-HSL present in every supernatant was evaluated (each reported fluorescence value was corrected with the O.D.600 of the culture). | ||
| Line 153: | Line 149: | ||
---- | ---- | ||
<br> | <br> | ||
| - | |||
===Microplate reader experiments for <partinfo>BBa_F2620</partinfo> - Protocol #4=== | ===Microplate reader experiments for <partinfo>BBa_F2620</partinfo> - Protocol #4=== | ||
| Line 168: | Line 163: | ||
<br> | <br> | ||
===Preliminary remarks=== | ===Preliminary remarks=== | ||
| - | *All our growth curves have been obtained subtracting for each time sample the broth O.D.600 measurement from that of the culture; broth was considered in the same conditions of the culture ( | + | *All our growth curves have been obtained subtracting for each time sample the broth O.D.600 measurement from that of the culture; broth was considered in the same conditions of the culture (i.e. induced with the same inducer concentration and supplemented with the same antibiotic of the culture). |
| - | *Fluorescence signals have been obtained subtracting for each time sample the fluorescent measurement of a non-fluorescent culture from that of the target culture. The non-fluorescent culture was considered in the same conditions of the | + | *Fluorescence signals have been obtained subtracting for each time sample the fluorescent measurement of a non-fluorescent culture from that of the target culture. The non-fluorescent culture was considered in the same conditions of the culture of interest (e.g. induced with the same inducer concentration and with the same plasmid/antibiotic resistance, but without fluorescent reporter genes). This operation allows the removal, from the target fluorescent signal, of the "self-fluorescent" component and the fluorescence signal obtained is "blanked". |
| Line 190: | Line 185: | ||
===Data analysis for self-inducible promoters (initiation-treshold determination)=== | ===Data analysis for self-inducible promoters (initiation-treshold determination)=== | ||
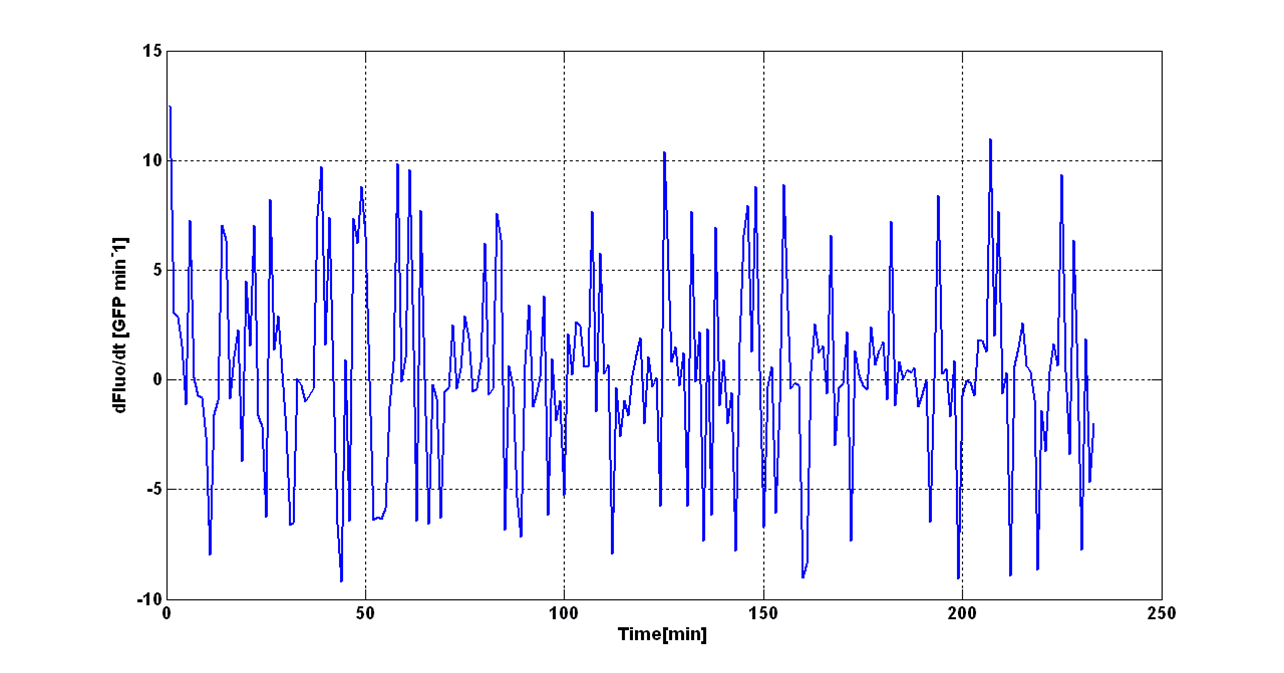
| - | The task of | + | The task of this analysis is the evaluation of the initiation transcription point (in terms of absorbance) for self-inducible devices. The transition O.D.600 value is named, from now on, O.D.start. |
| - | Data from three indipendent wells were averaged and blanked. O.D.600 signals were blanked as described in "Preliminary Remarks" section, while the fluorescence signal was blanked with the fluorescence of <partinfo>BBa_T9002</partinfo> part, assembled in the same plasmid of the considered promoter. This operation allows the removal from the fluorescence signal of the autofluorescence component and of the | + | Data from three indipendent wells were averaged and blanked. O.D.600 signals were blanked as described in "Preliminary Remarks" section, while the fluorescence signal was blanked with the fluorescence of <partinfo>BBa_T9002</partinfo> part, assembled in the same plasmid of the considered promoter. This operation allows the removal from the fluorescence signal of the autofluorescence component and of the “leaky” component (due to the leakage of ''lux pR'' promoter in absence of the autoinducer molecule HSL). |
| - | + | O.D.start was evaluated by computing the ''Scell'' (GFPmut3 synthesis rate per cell) signal for the desired self-inducible promoter. | |
| - | + | ||
| - | + | Scell was obtained by computing (1/O.D.600)*dGFP/dt, where O.D.600 and GFP are the blanked absorbance and fluorescence signals. | |
| - | + | The goal is the estimation of the critical O.D.600 value (O.D.start) at which the Scell significantly increases. Because Scell is a very noisy signal, a threshold value which takes into account the noise variability was proposed and it is described below. | |
| - | F_C1 and F_C2 have the same expected value and the same standard deviation, since they are two independent realizations of the same aleatory process: in fact, they are two time series acquired from the same cultures | + | Two different signals, measured from independent samples of the same non-fluorescent culture in the same experiment, are considered: C1 and C2. The fluorescence signal of C1 and C2 (F_C1 and F_C2 respectively) can be thought as the addition of a “real signal” and a noise component. |
| + | |||
| + | [[Image:UNIPV_Pavia_noise1.png|200px|center]] | ||
| + | |||
| + | F_C1 and F_C2 have the same expected value and the same standard deviation, since they are two independent realizations of the same aleatory process: in fact, they are two time series acquired from the same cultures in the same growth conditions by the same instrument. | ||
Noise signal was computed as: | Noise signal was computed as: | ||
[[Image:UNIPV_Pavia_noise2.png|150px|center]] | [[Image:UNIPV_Pavia_noise2.png|150px|center]] | ||
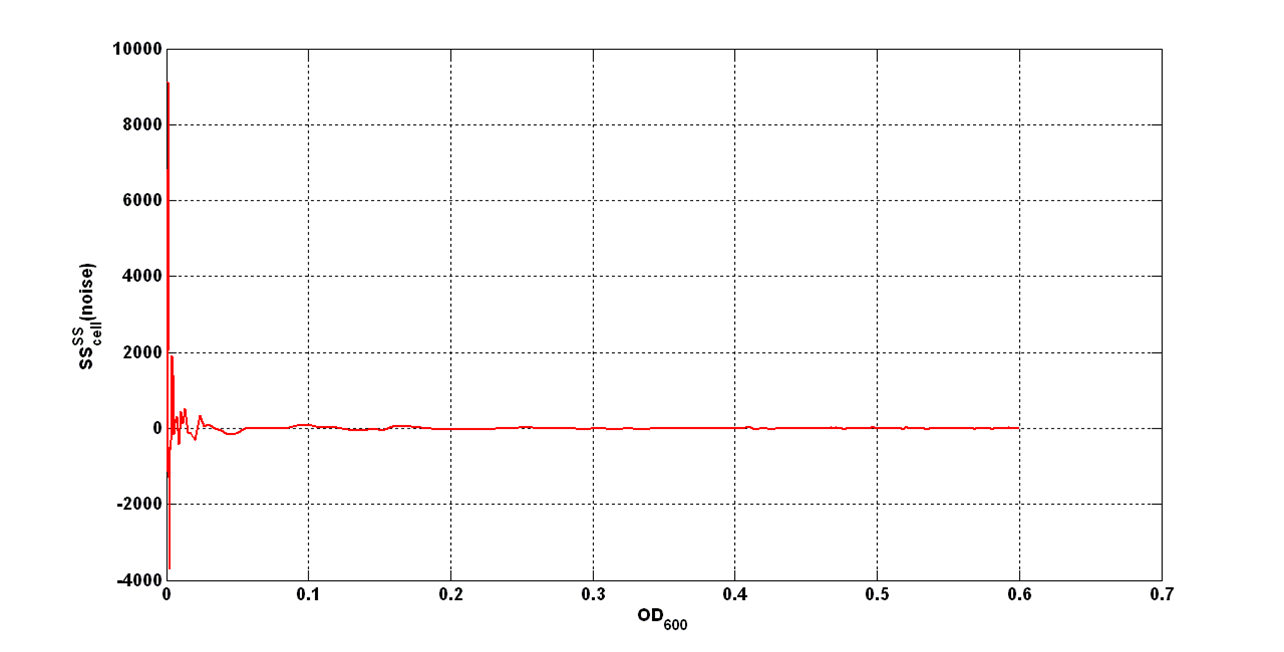
| - | + | An interesting signal is the Scell of ''N'' time series. It can be evaluated as the time derivative of ''N'', divided by O.D.600 of the culture. It has the behaviour shown in figure: | |
| - | + | ||
| - | + | ||
| - | An interesting signal is Scell of N. It can be evaluated as the time derivative of N, divided by O.D.600 of the culture. It has the behaviour shown in figure: | + | |
[[Image:UNIPV_Pavia_rumoreNoiseScell.png|600px|center|Scell Noise]] | [[Image:UNIPV_Pavia_rumoreNoiseScell.png|600px|center|Scell Noise]] | ||
| - | It is fair to say that the noise of Scell is bigger for low O.D.600 values and its | + | It is fair to say that the noise of Scell is bigger for low O.D.600 values and its amplitude decreases dramatically for higher O.D.600 values (e.g.: O.D.600>0.1 measured with the TECAN Infinite F200 in a 96-well microplate). |
| - | The noise model proposed for the Scell | + | The O.D.600-dependent noise model proposed for the Scell signal is: |
[[Image:UNIPV_Pavia_ModelNoiseScell.png|300px|center|Scell noise model]] | [[Image:UNIPV_Pavia_ModelNoiseScell.png|300px|center|Scell noise model]] | ||
| - | + | According to the formula reported above, it is possible to obtain the sigma_bar value as follows: | |
| + | #multiply the Scell time series values with the corresponding O.D.600; | ||
| + | #the sigma_bar constant value can be estimated as the standard deviation of this signal. | ||
| + | |||
| + | The Scell*O.D.600 signal behaviour is reported below: | ||
| + | |||
| + | [[Image:UNIPV_Pavia_rumoreNoise.png|600px|center|Measurement Noise]] | ||
| + | |||
| + | As this figure shows, the noise amplitude of this signal is no more O.D.600-dependent and the sigma_bar can be obtained. | ||
| + | |||
| + | |||
| + | The evaluation of the O.D.start can be performed supposing that the transcription starts when the signal is significantly different from the noise. A threshold on the signal amplitude was estabilished as reported here: | ||
[[Image:UNIPV_Pavia_threshold.png|300px|center|variance threshold]] | [[Image:UNIPV_Pavia_threshold.png|300px|center|variance threshold]] | ||
| + | |||
| + | In order to make the method more robust, induction was considered only when 5 consecutive values exceeded the threshold amplitude. | ||
| + | |||
The operation of subtracting the two signals F_C1 and F_C2 is analogous to the “blanking” operation performed on data, as described previously. For, this reason, under the hypothesis that the signals have the same variance (this is a consistent hypothesis, since they are measured by the same instrument in the same experimental conditions), this argument can be extended to the “blanked” data considered in the processing. | The operation of subtracting the two signals F_C1 and F_C2 is analogous to the “blanking” operation performed on data, as described previously. For, this reason, under the hypothesis that the signals have the same variance (this is a consistent hypothesis, since they are measured by the same instrument in the same experimental conditions), this argument can be extended to the “blanked” data considered in the processing. | ||
| - | In particular, it is evident that the "blanked" signal is affected by the same noise described above and thus, the noise of | + | In particular, it is evident that the "blanked" signal is affected by the same noise described above and thus, the noise of Scell signal is analogous to the one described before. |
Thus, this threshold was used to compute the O.D.start for the cultures. This heuristic algorithm was implemented in Matlab and analysis results are reported in the “results” section. | Thus, this threshold was used to compute the O.D.start for the cultures. This heuristic algorithm was implemented in Matlab and analysis results are reported in the “results” section. | ||
| - | |||
| - | |||
| Line 234: | Line 241: | ||
<br> | <br> | ||
| - | + | ===Data analysis to estimate the HSL synthesis rate per cell=== | |
| - | ===Data | + | |
The autoinducer synthesys rate per cell is a very important parameter to be evaluated in quorum sensing systems. | The autoinducer synthesys rate per cell is a very important parameter to be evaluated in quorum sensing systems. | ||
| - | In fact the knowledge of this parameter enables the rational design of cell-communication systems, such as the self inducible devices studied in this project. | + | In fact the knowledge of this parameter enables the rational design of cell-communication systems, such as the self-inducible devices studied in this project. |
A model based approach was proposed to estimate this interesting parameter. | A model based approach was proposed to estimate this interesting parameter. | ||
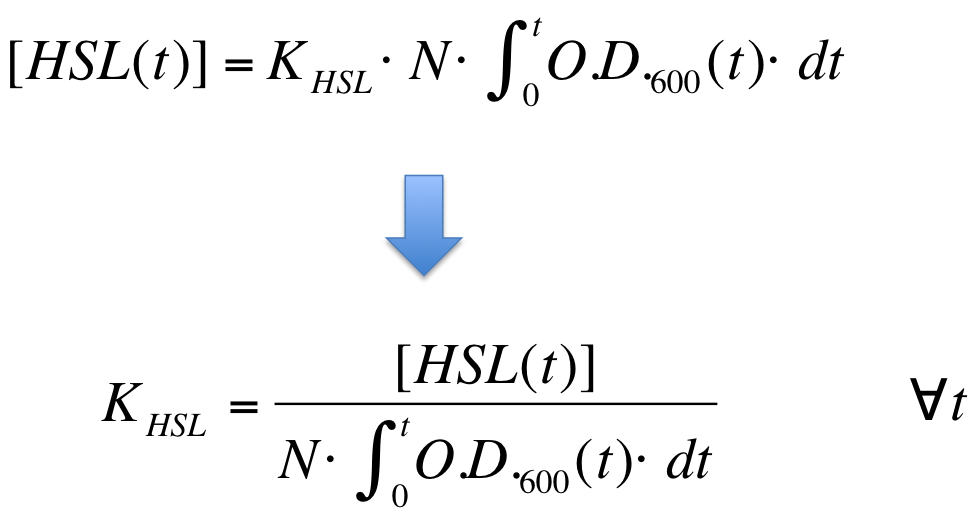
| Line 263: | Line 269: | ||
|[[Image:PV_Immagine_formula2.png|300px]] | |[[Image:PV_Immagine_formula2.png|300px]] | ||
|} | |} | ||
| - | K_HSL can be estimated for t=t_bar, corresponding to the | + | K_HSL can be estimated for t=t_bar, corresponding to the transcription initiation time, as reported below. |
{| align='center' | {| align='center' | ||
| Line 270: | Line 276: | ||
where: | where: | ||
* N was estimated by counting TOP10 colonies obtained by plating serial dilutions of cultures with known O.D.600. | * N was estimated by counting TOP10 colonies obtained by plating serial dilutions of cultures with known O.D.600. | ||
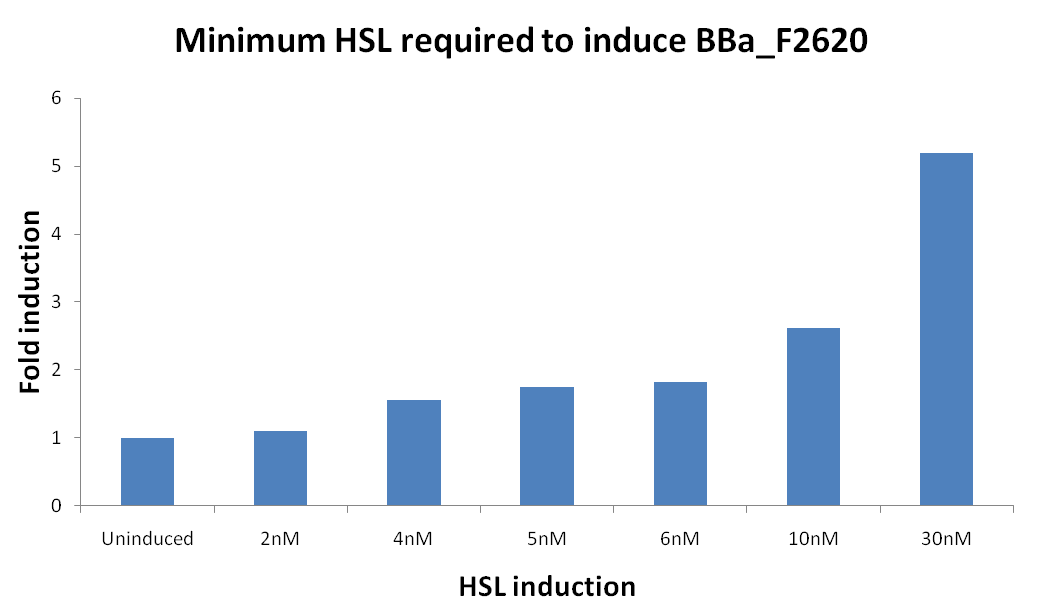
| - | * [HSL(t_bar)] was estimated as described in the section [[Data Analysis - minimum induction required to activate ''lux pR'' for <partinfo>BBa_F2620</partinfo>]] | + | * [HSL(t_bar)] was estimated as described in the section [[Team:UNIPV-Pavia/Parts/Characterization#Data Analysis - minimum induction required to activate lux pR for BBa_F2620|Data analysis - minimum induction required to activate ''lux pR'' for <partinfo>BBa_F2620</partinfo>]] |
* the O.D.600 time series was measured in each experiment, so its time integral can be computed by trapezoidal numerical integration. | * the O.D.600 time series was measured in each experiment, so its time integral can be computed by trapezoidal numerical integration. | ||
| - | N was estimated and values are reported below: | + | N was estimated in LB and M9 media and values are reported below: |
{| border='1' align='center' | {| border='1' align='center' | ||
| Line 312: | Line 318: | ||
===Data analysis for RPU evaluation=== | ===Data analysis for RPU evaluation=== | ||
| - | The RPUs are standard units proposed by Kelly J. et al., | + | The RPUs are standard units proposed by Kelly J. et al., 2009, in which the relative transcriptional strength of a promoter can be measured using a reference standard. |
RPUs have been computed as: | RPUs have been computed as: | ||
Latest revision as of 02:42, 28 October 2010
|
|
|||||||||||||||||||||||||
|
|
||||||||||||||||||||||||
 "
"