Team:Uppsala-Sweden/Sponsors
From 2010.igem.org
(→Acknowledgement) |
(→Acknowledgement) |
||
| (10 intermediate revisions not shown) | |||
| Line 25: | Line 25: | ||
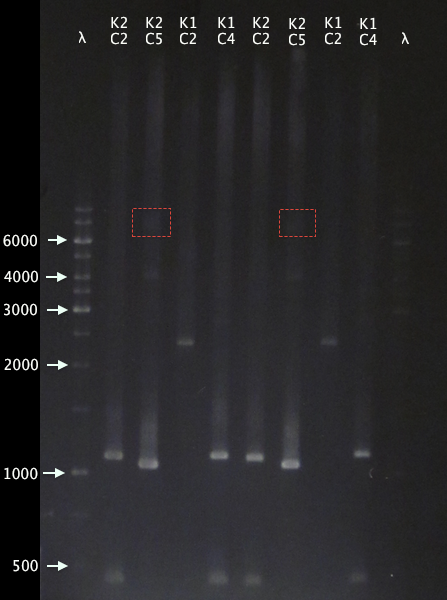
We provided the Stockholm team, represented by Nina Schiller,our K1and K2 constructs and their result are showed below. | We provided the Stockholm team, represented by Nina Schiller,our K1and K2 constructs and their result are showed below. | ||
| - | [[Image:Uppsala Stockholm.png| | + | [[Image:Uppsala Stockholm.png|400px|border|left|thumb|Fig 1]] |
| - | [[Image:Uppsala Stockholm2.png| | + | [[Image:Uppsala Stockholm2.png|445px|border|right|thumb|Fig 2]] |
| + | |||
| + | |||
| + | |||
| + | . | ||
| + | I | ||
Fig.1 shows our construct the gel run of the K1 and K2 PCR-amplified samples The red-marked rectangulars is the expected length of the K1 and K2 constructs while the bands only reach slighty above the 1000bp marker. One possible explanation the Stockholm team brought up, was the fact that the primers VF2 and VR have a tendency to mispriming with BB_R0062 and BB_B0010 respectively when performing a PCR, http://partsregistry.org/Problems_with_PCR_using_VR/VF2. Our K- construct contains BB_F2621 which has the LuxR promoter BB_R0062 and severeal BB_B0015 termintors which has the BB_B0010. | Fig.1 shows our construct the gel run of the K1 and K2 PCR-amplified samples The red-marked rectangulars is the expected length of the K1 and K2 constructs while the bands only reach slighty above the 1000bp marker. One possible explanation the Stockholm team brought up, was the fact that the primers VF2 and VR have a tendency to mispriming with BB_R0062 and BB_B0010 respectively when performing a PCR, http://partsregistry.org/Problems_with_PCR_using_VR/VF2. Our K- construct contains BB_F2621 which has the LuxR promoter BB_R0062 and severeal BB_B0015 termintors which has the BB_B0010. | ||
Latest revision as of 10:55, 27 October 2010



Sponsors
Heartfelt thanks to the Technical Faculty and IBG (Institutionen för biologisk grundutbildning) department of Uppsala University, for supporting our team.
Our sincere thanks to Prof. Peter Lindblad of the Department of Photochemistry and Molecular Science, Uppsala University for giving us the space and resources to execute our project in his lab.
We would like to thank Inger Jonasson, Uppsala Genome Center Dept of Genetics and Pathology, Rudbeck Laboratory, Uppsala University for help with confirming the constructed plasmids.
Thanks to [http://www.genscript.com/ GenScript] for sponsoring our Tshirts
Acknowledgement
Stockholm iGEM team
We want to acknowledge the Stockholm iGEM team for contributing to our work. We provided the Stockholm team, represented by Nina Schiller,our K1and K2 constructs and their result are showed below.
. I
Fig.1 shows our construct the gel run of the K1 and K2 PCR-amplified samples The red-marked rectangulars is the expected length of the K1 and K2 constructs while the bands only reach slighty above the 1000bp marker. One possible explanation the Stockholm team brought up, was the fact that the primers VF2 and VR have a tendency to mispriming with BB_R0062 and BB_B0010 respectively when performing a PCR, http://partsregistry.org/Problems_with_PCR_using_VR/VF2. Our K- construct contains BB_F2621 which has the LuxR promoter BB_R0062 and severeal BB_B0015 termintors which has the BB_B0010. A closer look shows band around 2500 bp for K1C2 and and some very pale ones for K2C5 with brighter unspecific bands.
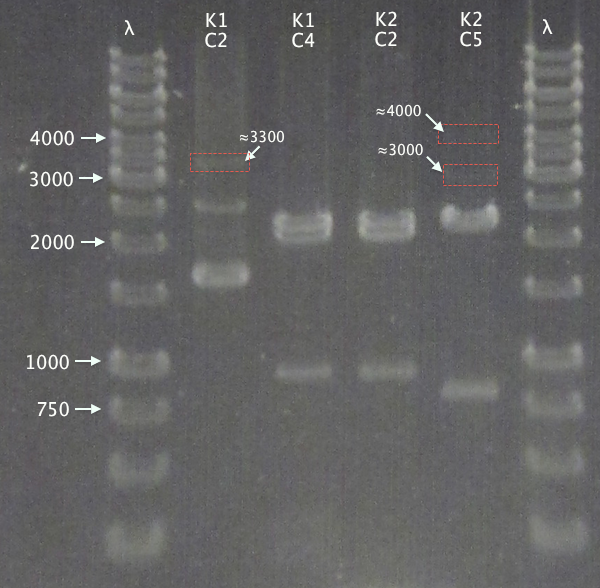
Fig2 shows the K constructs digested with EcorI and Pst1.The expected length is around 6-7 Kbp but as in Fig.1, there are bands around 1000 bp. Further up hicker bands appears around 1500-2500 bp and according to the StockholmTeam, these are the result of uncompleted digestion by the restriction enzymes
Department of Photochemistry and Molecular Science.
We also want to thank the department of Photochemistry and Molecular Science in Uppsala, specially Professor Lindblad for providing us access to his lab and letting us use the equipment and resources necesseary for our project. We also want to thank our supervisors Thorsten Heidorn, Daniel Camsund and Hsin-Ho Huang for supporting and giving us advice an tips during the project.
Uppsala University
We want to thank the Technical Faculty and the Biological Education Centre so much for the financial support making the team´s attendance at the Jamboree in MIT, possible. We really appreciate that they have done for us, specially considering that the team took contact with both the university organisations quite late into the iGEM- competition.
GenScript
A big also to GenScript for providing us with our cool T-shirts (Fig3) with our own logo and a cash coupon of US$500 for purchase gene synthesis service that can be used in other research projects.
 "
"