Team:Imperial College London/Modelling/Output/Detailed Description
From 2010.igem.org
m |
m |
||
| (14 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
{{:Team:Imperial_College_London/Templates/Buttons}} | {{:Team:Imperial_College_London/Templates/Buttons}} | ||
| - | {{:Team:Imperial_College_London/Templates/ | + | {{:Team:Imperial_College_London/Templates/ModellingHeaderF}} |
{{:Team:Imperial_College_London/Templates/ModellingOutputHeader}} | {{:Team:Imperial_College_London/Templates/ModellingOutputHeader}} | ||
| - | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top: | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" |
|style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Detailed Description | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Detailed Description | ||
|- | |- | ||
| Line 13: | Line 13: | ||
<li>Enzymatic reactions</li> | <li>Enzymatic reactions</li> | ||
</ol> | </ol> | ||
| - | + | |} | |
| - | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | |
| - | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|1. Law of Mass Action | |
| + | |- | ||
| + | | | ||
During a meeting with our advisors, it was noted that our initial models (in which it was assumed that our system obeyed Michaelis-Menten kinetics) were wrong as the assumptions made by Michaelis-Menten approximation were not obeyed by the system. | During a meeting with our advisors, it was noted that our initial models (in which it was assumed that our system obeyed Michaelis-Menten kinetics) were wrong as the assumptions made by Michaelis-Menten approximation were not obeyed by the system. | ||
| Line 177: | Line 179: | ||
As we could not use the Michaelis-Menten simplification to model enzymatic reactions in our system, we will had to solve the problem from first principle. It meant referring to more general set of assumptions called Law of Mass Action. This allowed us to model our enzymatic reactions without making assumptions about amounts of particular species as long as the amounts are bigger than single molecule level. This resulted in bigger number of partial differential equations as there was one per each species instead of 1 per reaction. | As we could not use the Michaelis-Menten simplification to model enzymatic reactions in our system, we will had to solve the problem from first principle. It meant referring to more general set of assumptions called Law of Mass Action. This allowed us to model our enzymatic reactions without making assumptions about amounts of particular species as long as the amounts are bigger than single molecule level. This resulted in bigger number of partial differential equations as there was one per each species instead of 1 per reaction. | ||
| - | + | |} | |
| - | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | |
| - | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|2. Model preA: Simple production of dioxygenase | |
| + | |- | ||
| + | | | ||
This model was developed to illustrate 1-step amplification output. | This model was developed to illustrate 1-step amplification output. | ||
| Line 202: | Line 206: | ||
<CENTER><img src="https://static.igem.org/mediawiki/2010/d/d3/IC_Equation_3.png"/></CENTER> | <CENTER><img src="https://static.igem.org/mediawiki/2010/d/d3/IC_Equation_3.png"/></CENTER> | ||
</html> | </html> | ||
| + | In the equation above: [p] = protein concentration, s = protein production rate, d = protein degradation rate | ||
The enzymatic interactions can be described in the following way: | The enzymatic interactions can be described in the following way: | ||
<html> | <html> | ||
| - | <CENTER><img src="https://static.igem.org/mediawiki/2010/ | + | <CENTER><img src="https://static.igem.org/mediawiki/2010/7/7b/IC_Equation_5.png"/></CENTER> |
</html> | </html> | ||
| - | + | In the equation above : D is Dioxygenase, C is Catechol, C-D is catechol-dioxygenase transition complex, M<sub>a</sub> is Muconic acid - the yellow ouptput. | |
It is important to mention dioxygenase molecules tretramerize before becoming active enzymes. It has been very simplified in the model. Tetramerization is accounted for by simply dividing dioxygenase concentration by 4 before it acts on catechol. | It is important to mention dioxygenase molecules tretramerize before becoming active enzymes. It has been very simplified in the model. Tetramerization is accounted for by simply dividing dioxygenase concentration by 4 before it acts on catechol. | ||
| Line 214: | Line 219: | ||
<b>Differential equations</b> | <b>Differential equations</b> | ||
| - | The above reaction can be written in terms of differential equations: | + | The above reaction can be written in terms of ordinary differential equations: |
<html> | <html> | ||
| - | <CENTER><img src=""/></CENTER> | + | <CENTER><img src="https://static.igem.org/mediawiki/2010/d/d7/IC_Equation_2.png"/></CENTER> |
</html> | </html> | ||
| Line 223: | Line 228: | ||
These equations were implemented in Matlab, using a built-in function (ode15s) which solves ordinary differential equations. For the code please refer to the <html><a href="https://2010.igem.org/Team:Imperial_College_London/Modelling/Output/Download_MatLab_Files"><b>Download MatLab Files</b></a></html> section. | These equations were implemented in Matlab, using a built-in function (ode15s) which solves ordinary differential equations. For the code please refer to the <html><a href="https://2010.igem.org/Team:Imperial_College_London/Modelling/Output/Download_MatLab_Files"><b>Download MatLab Files</b></a></html> section. | ||
| - | + | |} | |
| - | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | |
| - | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|3. Model A: Activation of Dioxygenase by TEV enzyme | |
| + | |- | ||
| + | | | ||
This model was developed to illustrate 1-step amplification output. | This model was developed to illustrate 1-step amplification output. | ||
| Line 234: | Line 241: | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
| - | {| style="background:#e1e1e1;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | + | {| style="width:704px;background:#e1e1e1;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; |
|- | |- | ||
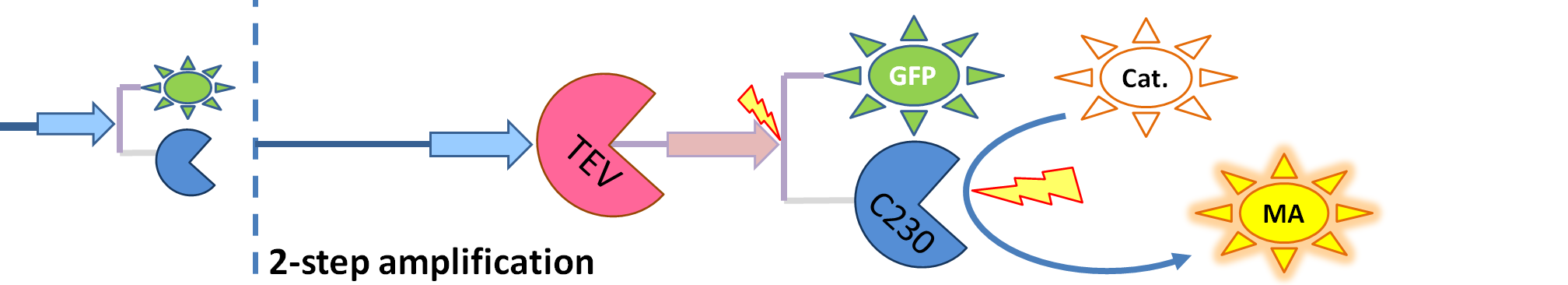
|[[Image:2-step_amplification.png|700px]] | |[[Image:2-step_amplification.png|700px]] | ||
| Line 245: | Line 252: | ||
This model includes 2 enzymatic reactions: | This model includes 2 enzymatic reactions: | ||
| - | + | <ol> | |
| - | + | <li>Reaction between dioxygenase and catechol, which is exactly the same as in model preA<br/> | |
| - | + | <html> | |
| - | <br/><html> | + | <CENTER><img src="https://static.igem.org/mediawiki/2010/7/7b/IC_Equation_5.png"/></CENTER> |
| - | <CENTER><img src="https://static.igem.org/mediawiki/2010/ | + | </html><br/> |
| + | In the equation above : D is Dioxygenase, C is Catechol, C-D is catechol-dioxygenase transition complex, M<sub>a</sub> is Muconic acid - the yellow ouptput.</li> | ||
| + | <li>Reaction between TEV and inactive dioxygenase<br/> | ||
| + | <html> | ||
| + | <CENTER><img src="https://static.igem.org/mediawiki/2010/6/68/IC_Equation_7.png"alt="Equations showing enzymatic reaction between TEV and split Dioxygenase" /></CENTER> | ||
</html> | </html> | ||
| + | <br/> | ||
| + | In the equation above: TEV = Protease of the Tobacco Etch Virus, sD = split Dioxygenase, TsD = intermediate complex of the enzymatic reaction,<br/>D = Dioxygenase</li> | ||
| + | </ol> | ||
<b>Differential equations</b> | <b>Differential equations</b> | ||
| Line 275: | Line 289: | ||
|} | |} | ||
</div> | </div> | ||
| - | + | |} | |
| - | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | |
| - | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|4. Model B: Activation of Dioxygenase by TEV or activated split TEV | |
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|enzyme | ||
| + | |- | ||
| + | | | ||
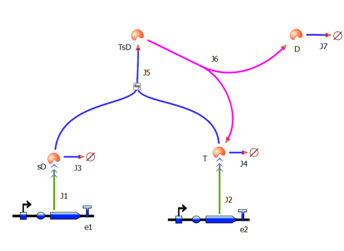
<b>Elements of the system</b> | <b>Elements of the system</b> | ||
# split TEV protease (shown <i>purple</i>) is introduced as the 3rd amplification step. It exists in 2 versions (not shown on diagram): a and b as it does not separate into 2 identical halves. | # split TEV protease (shown <i>purple</i>) is introduced as the 3rd amplification step. It exists in 2 versions (not shown on diagram): a and b as it does not separate into 2 identical halves. | ||
| Line 305: | Line 323: | ||
<html> | <html> | ||
| - | <CENTER><img src="https://static.igem.org/mediawiki/2010/ | + | <CENTER><img src="https://static.igem.org/mediawiki/2010/c/c6/IC_Equation_8.png"/></CENTER> |
</html> | </html> | ||
| + | In the equation above: T = Tobacco Etch Virus Protease, sD = split Dioxygenase, D = Dioxygenase, sTa/sTb = inactivated split TEV, Ts/Tsa/Tsb = activated split TEV | ||
<b>Implementation in Matlab</b> | <b>Implementation in Matlab</b> | ||
For the code please refer to the <html><a href="https://2010.igem.org/Team:Imperial_College_London/Modelling/Output/Download_MatLab_Files"><b>Download MatLab Files</b></a></html> section. | For the code please refer to the <html><a href="https://2010.igem.org/Team:Imperial_College_London/Modelling/Output/Download_MatLab_Files"><b>Download MatLab Files</b></a></html> section. | ||
| - | + | |} | |
| - | + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | |
| - | + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|5. Model C: Further improvements | |
| + | |- | ||
| + | | | ||
This model has not been implemented because of the conclusions that we reached from Models A and B. | This model has not been implemented because of the conclusions that we reached from Models A and B. | ||
It would include the following features: | It would include the following features: | ||
*activated split TEV (TEVs) is allowed to activate not only sD but sTEVa and sTEVb | *activated split TEV (TEVs) is allowed to activate not only sD but sTEVa and sTEVb | ||
| + | |- | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;" align="right"|[[Team:Imperial_College_London/Modelling/Output/Results_and_Conclusion | Click here for the results of this model...]] | ||
|} | |} | ||
Latest revision as of 03:48, 28 October 2010
| Modelling | Overview | Detection Model | Signaling Model | Fast Response Model | Interactions |
| A major part of the project consisted of modelling each module. This enabled us to decide which ideas we should implement. Look at the Fast Response page for a great example of how modelling has made a major impact on our design! | |
| Objectives | Description | Results | Constants | MATLAB Code |
| Detailed Description |
|
The following page presents the details of the models that have been developed. Firstly, assumptions that have been exploited are explained. Then every model is presented separately as each of them has slightly different elements of the system and the interactions between them. However, there are only 3 fundamental biochemical processes that will be analysed:
|
| 1. Law of Mass Action | ||||||||||||||||||||||||||||
|
During a meeting with our advisors, it was noted that our initial models (in which it was assumed that our system obeyed Michaelis-Menten kinetics) were wrong as the assumptions made by Michaelis-Menten approximation were not obeyed by the system. A few of Michaelis-Menten assumptions were not met by our system:
Since we are continuously producing enzyme, Vmax will change. Therefore the conservation of enzymes E0 = E + ES does not hold for our system.
We are producing both substrate and enzyme, so we have approximately the same amount of substrate and enzyme.
Abandoned Initial Attempts
Elements of the system
Simple models
Implementation of Michaelis-Menten kinetics
Conclusion We were not able to obtain all the necessary constants. Hence, we decided to make educated guesses about possible relative values between the constants as well as varying them and observing the change in output. As the result, we concluded that the amplification happens at each amplification level proposed. The magnitude of amplification varies depending on the constants. There is not much difference between using TEV or HIV1. Change of output During our literature research, we came across a better output, so we abandoned the idea of using GFP as an output. Instead, we are using catechol. An enzyme, dioxygenase, will be acting on the catechol, which will then result in a coloured output. Catechol will be added to the bacteria manually (i.e. the bacteria will not produce catechol). Hence, in our models dioxygenase will be treated as an output as this enzyme is the only activator of catechol in our system. This means that the change of catechol into its colourful form is dependent on the dioxygenase concentration. References
As we could not use the Michaelis-Menten simplification to model enzymatic reactions in our system, we will had to solve the problem from first principle. It meant referring to more general set of assumptions called Law of Mass Action. This allowed us to model our enzymatic reactions without making assumptions about amounts of particular species as long as the amounts are bigger than single molecule level. This resulted in bigger number of partial differential equations as there was one per each species instead of 1 per reaction. |
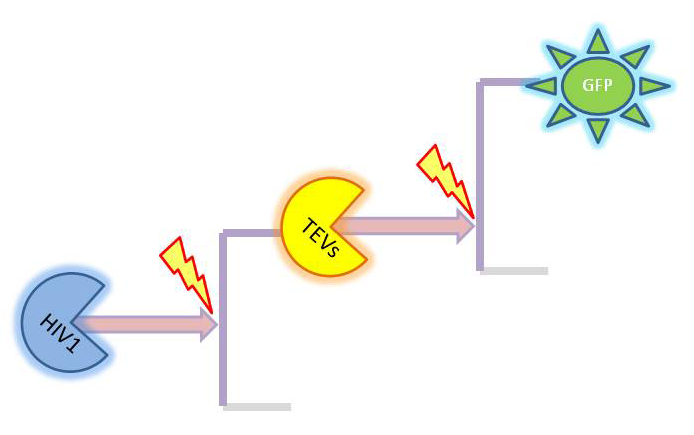
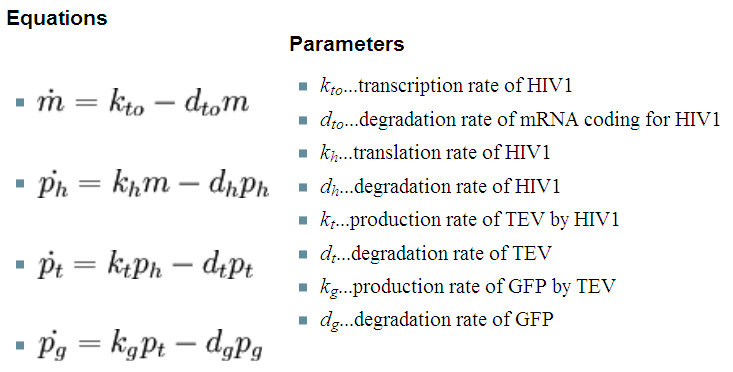
| 2. Model preA: Simple production of dioxygenase | ||
|
This model was developed to illustrate 1-step amplification output. Elements of the system
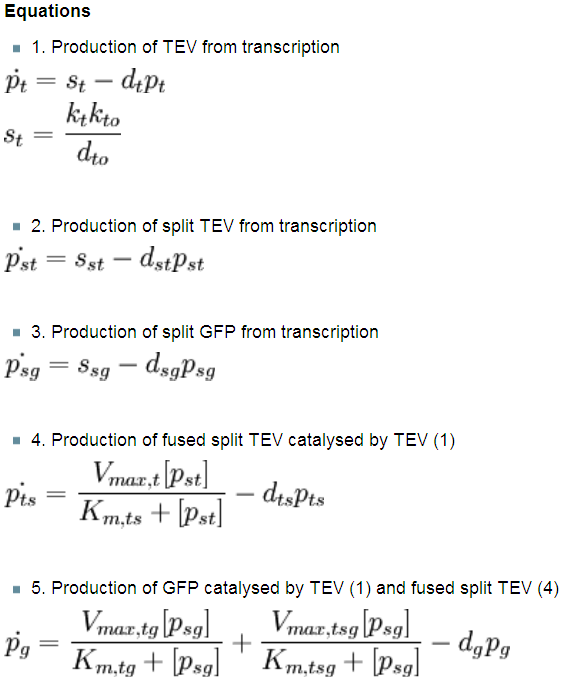
Interactions between elements Initially, dioxygenase has to be transcribed and translated. This has been described using 1 ODE equation:
 The enzymatic interactions can be described in the following way:
 It is important to mention dioxygenase molecules tretramerize before becoming active enzymes. It has been very simplified in the model. Tetramerization is accounted for by simply dividing dioxygenase concentration by 4 before it acts on catechol. Differential equations The above reaction can be written in terms of ordinary differential equations:
 Implementation in Matlab These equations were implemented in Matlab, using a built-in function (ode15s) which solves ordinary differential equations. For the code please refer to the Download MatLab Files section. |
| 3. Model A: Activation of Dioxygenase by TEV enzyme |
|
This model was developed to illustrate 1-step amplification output. Elements of the system
Interactions between elements This model includes 2 enzymatic reactions:
Differential equations This is a simple enzymatic reaction, where TEV is the enzyme, Dioxygenase the product and split Dioxygenase the substrate. Choosing k1, k2, k3 as reaction constants, the reaction can be rewritten in these four sub-equations:
 Implementation in Matlab These equations were implemented in Matlab, using a built-in function (ode15s) which solves ordinary differential equations. For the code please refer to the Download MatLab Files section. Implementation in TinkerCell Another approach to model the amplification module would be to implement it in a program such as TinkerCell (or CellDesigner). This was used to check whether implementation in Matlab generates similar results. If happened otherwise, we would need to look for reasons for those differences in the programs. As the results, generated by Matlab were the same, only Matlab code has been developed further as it allows more flexibility, control and insight. |
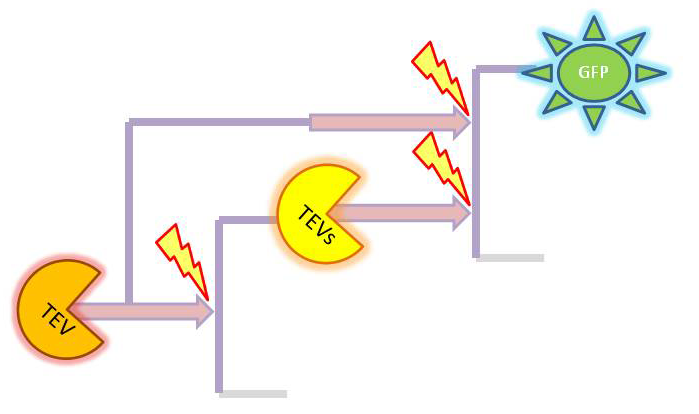
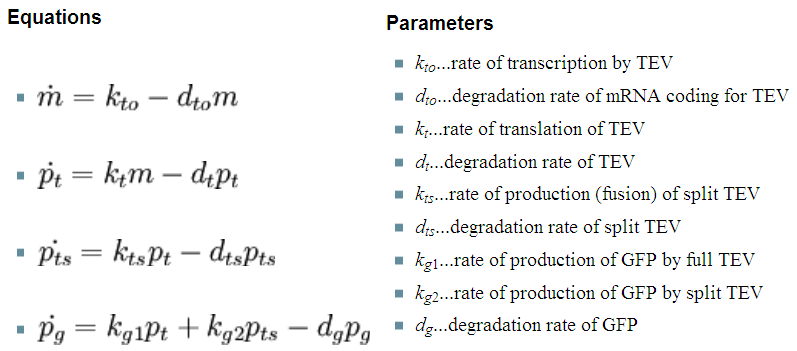
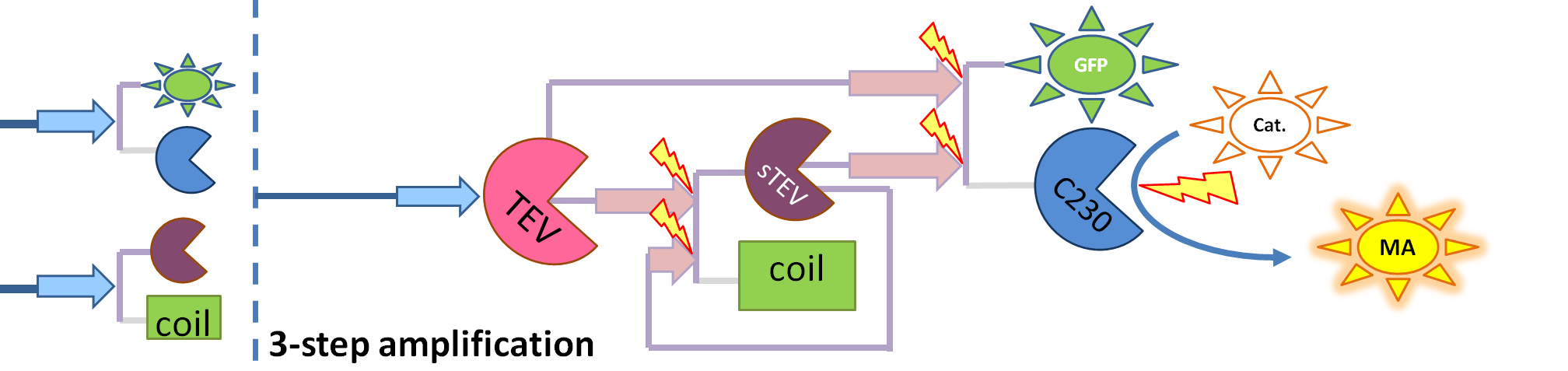
| 4. Model B: Activation of Dioxygenase by TEV or activated split TEV |
| enzyme |
|
Elements of the system
Interactions and assumptions This version includes the following features:
 Implementation in Matlab For the code please refer to the Download MatLab Files section. |
| 5. Model C: Further improvements |
|
This model has not been implemented because of the conclusions that we reached from Models A and B. It would include the following features:
|
| Click here for the results of this model... |
 "
"