Team:Heidelberg/Modeling
From 2010.igem.org
(→Input/target pairs) |
AlejandroHD (Talk | contribs) (→Modeling approach to the project) |
||
| (51 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| + | {{:Team:Heidelberg/Single_Pagetop|mode_over}} | ||
| + | {{:Team:Heidelberg/Side_Top}} | ||
| - | + | __TOC__ | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | {{:Team:Heidelberg/Side_Bottom}} | |
| + | =Modeling approach to the project= | ||
| + | As the title of our project states, '''“DNA is not enough”'''. There are several upper-level regulation systems in higher organisms. Our main idea was to use one of them to tune the expression of genes and device a tissue-specific gene therapy approach. | ||
| + | The miBricks project consists basically of two ideas. The first is tuning of gene expression using shRNAs/miRNAs and the second is specific targeting of tissues. We intend to tune the expression of a gene by manipulating the binding affinity of a miRNA/shRNA towards the transcript of this gene which results in different expression levels. In order to do this, different binding sites for the miRNA/shRNA are introduced in the 3'UTR of the gene of interest. These binding sites differ from each other in terms of certain sequence-based features. By computational methods, we predict the binding site that should be inserted to achieve the level of expression desired. Targeting of specific tissues is achieved by introducing binding sites for tissue-specific endogenously-expressed miRNA into the construct, thus causing knockdown of a gene based on its presence or absence in the tissue. | ||
| - | + | Apart from several bioinformatic tools, our team developed two independent models: | |
| - | The | + | The [https://2010.igem.org/Team:Heidelberg/Modeling/trainingset#Neural_Network_Model Neural Network Model] takes inspiration in the biological nervous system to predict its results. It is the appropriate strategy to model complex processes and it is able to learn from experience. Neural Networks generally require a big amount of data to be fully trained. Even though the experimental data was limiting, the results agree with the experimental values and the model was able to determine the importance of the bulge size for knockdown. |
| - | + | The [https://2010.igem.org/Team:Heidelberg/Modeling/trainingset#Fuzzy_Logic_Model Fuzzy Logic Model] is combining the strength of intuitive integration of prior knowledge with a sophisticated Global Genetic optimization Algorithm. After training the model, it was able to reproduce the experimental data, especially the correlation in a 3-dimensional space of the AU content score and 3' pairing score to the knockdown percentage. | |
| + | <br> | ||
| + | <br> | ||
| + | To create our informatic tool to support the project, we looked at the problem from three different perspectives: | ||
| - | + | - Adjustment of gene expression in specific tissues. | |
| + | |||
| + | - Tuning expression level accurately. | ||
| - | + | - Predefine a construct to be used. | |
| - | + | ||
| - | + | ||
| - | + | These three paths were the inspiration behind creating miBEAT (miRNA Binding Site Engineering and Assembly Tool), to provide a strategy which makes possible to control the expression of genes in a specific way between tissues. | |
| - | + | To make this work, we tried to match the functionalities of the tool to our experimental project. Additionally, we provided a strategy that guides the user through the cloning process and allows them to use characterized standard parts sent to the MIT parts registry. | |
| - | + | ||
| - | + | Our complete work is present in the form of a graphical user interface called [https://2010.igem.org/Team:Heidelberg/Modeling/miGUI miBEAT]. This tool combines and connects the output of the different models and scripts and then generates a suitable miTuner construct that will express the gene of interest, miGENE, up or down to the desired level. miBEAT consists of three subparts; miRockdown, miBS designer and mUTING. | |
| - | + | ||
| - | + | ||
| - | + | [https://2010.igem.org/Team:Heidelberg/Modeling/miRockdown miRockdown] is the subpart which contains two computational models that work on different concepts: Neural Network and Fuzzy Logic plus the experimentally obtained data. | |
| - | [ | + | The models are sequentially associated with a script based on [http://www.targetscan.org/ Target Scan] algorithm. miRockdown takes as an input the desired knockdown percentage and the sequence of shRNAmir and gives out binding site parameters that are then compared with model predictions to finally generate the appropriate binding site. |
| - | + | [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner miBS designer] is available as a stand alone for generating customized binding sites, but a modified version of it is also a part of miBEAT, in charge of generating more than 2000 different binding sites for every miRNA sequences, following more than 135 combinations of regions. | |
| + | [https://2010.igem.org/Team:Heidelberg/Modeling/miSpec mUTING] provides the tissue specific targeting function to the GUI. It uses literature data for miRNA expression in various tissues and can output miRNA binding sites that could be used to differentiate between target and off target tissues. | ||
| - | + | =miRNA binding site features= | |
| - | - | + | miRNA are non-coding regulatory RNAs functioning as post-transcriptional gene silencers. After they are processed, they are usually 22 nucleotides long and they usually bind to the 3’UTR region of the mRNA (although they can also bind to the ORF or to the 5'UTR), forcing the mRNA into degradation or just repressing translation [Bartel, 2004]. |
| - | + | In vegetal organisms, miRNA usually bind to the mRNA with extensive complementarity. In animals, interactions are more inexact, creating a lot of uncertainty in the in silico prediction of targets[?]. | |
| - | + | The seed of the miRNA is usually defined as the region centered in the nucleotides 2-7 in the 5’ end of the miRNA. For an efficient binding site extensive pairing is usually required between the seed and the corresponding part of the mRNA. The seed, and the corresponding pairing sequence of the mRNA are located inside the AGO protein. | |
| - | + | Common types of miRNA seeds: | |
| - | - | + | - 6mer (abundance 21.5%): only the nucleotides 2-7 of the miRNA match with the mRNA. |
| - | + | - 7merA1 (abundance 15.1%): the nucleotides 2-7 match with the mRNA, and there is an adenine in position 1. | |
| - | + | - 7merm8 (abundance 25%): the nucleotides 2-8 match with the mRNA. | |
| - | + | ||
| - | + | - 8mer (abundance 19.8%): the nucleotides 2-8 match with the mRNA and there is an adenine in position 1. | |
| - | + | ||
| - | + | The percentages of abundance are calculated among conserved mammalian sites for a highly conserved miRNA (Friedman et al. 2008) | |
| - | + | ||
| - | + | <center>[[Image:Final_sequences_miRNAseeds.png|800 px]]<br> | |
| - | + | ||
| + | Figure 1: Interactions between two miRNAs and their binding sites. Notice the different types of seeds.</center> | ||
| - | + | Outside the seed, the existence of supplemental pairing (at least 3 contiguous nucleotides and at best centered in nucleotides 13-16 of the miRNA) stabilizes the bound complex and increases the efficacy of the binding site. | |
| - | + | ||
| - | + | ||
| - | + | Binding sites with a high local AU content around the binding site have proven to be more effective (possibly because of the destabilization of the mRNA secondary structure around the site). | |
| - | + | An arginine at position one of the binding site supposedly binds to a different protein of the RISC complex [Bartel, 2009], thus increasing the binding site efficiency significantly. | |
| - | + | Binding sites at the end or the beginning of the 3'UTR are more efficient. Binding sites within the first 15 nucleotides after the stop codon are not effective, since this region of the mRNA is inside the ribosome when translations stops. Thus a bound RISC complex in this region will dissociate after every round of translation and can not follow it's usual mode of action [Grimson et al., 2007]. | |
| - | |||
| - | == | + | =Tissue specific miRNAs= |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | A useful supplement to achieving tuned gene expression in cells is the ability to specifically target tissues where this should be carried out. Tissue targeting has, for quite some time, been an important field of research that has drawn much attention and is central to gene therapy. [https://2010.igem.org/Team:Heidelberg/Modeling/miGUI miBEAT] tool not only allows generation of binding sites that regulate the level of expression of a desired gene, but also employs strategies that help target the right tissue and exclude expression in others. This functionality is based on the principle of using tissue specific miRNA binding sites which can be introduced in the [https://2010.igem.org/Team:Heidelberg/Project/miRNA_Kit miTuner] construct easily. | |
| - | + | A smart way to specifically target tissues is to exploit the presence and absence of tissue specific endogenous miRNA in the target to specifically express or exclude expression of the gene of interest in the target. We make use of two of strategies based on this principle, namely, on-targeting and off-targeting. The off-targeting concept has been applied previously {{HDref|Wenfang Shi et.al., 2008}} wherein an endogenous miRNA is selected such that it is not present in the target tissue (therefore the gene is expressed) and is present in all the off target tissues (knockdown of the transcripts). Thus the gene is specifically expressed in the target. | |
| - | + | ||
| - | + | In addition to the off-targeting strategy, we designed a new strategy, the on-targeting. In this case, the miRNA is present in the target tissue and excluded from the off-targets. The binding site for this miRNA is present within the 3'UTR of a repressor gene (in our case TET/O2) construct. The operator for the repressor in turn precedes the gene of interest (miGene) in the miTuner or miMeasure constructs. Therefore in the presence of miRNA in the cell, repressor is degraded and miGene is expressed while in off-targets repressor is translated and represses the expression of miGene. | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Bottom}} |
Latest revision as of 03:48, 28 October 2010

Modeling approach to the projectAs the title of our project states, “DNA is not enough”. There are several upper-level regulation systems in higher organisms. Our main idea was to use one of them to tune the expression of genes and device a tissue-specific gene therapy approach. The miBricks project consists basically of two ideas. The first is tuning of gene expression using shRNAs/miRNAs and the second is specific targeting of tissues. We intend to tune the expression of a gene by manipulating the binding affinity of a miRNA/shRNA towards the transcript of this gene which results in different expression levels. In order to do this, different binding sites for the miRNA/shRNA are introduced in the 3'UTR of the gene of interest. These binding sites differ from each other in terms of certain sequence-based features. By computational methods, we predict the binding site that should be inserted to achieve the level of expression desired. Targeting of specific tissues is achieved by introducing binding sites for tissue-specific endogenously-expressed miRNA into the construct, thus causing knockdown of a gene based on its presence or absence in the tissue. Apart from several bioinformatic tools, our team developed two independent models: The Neural Network Model takes inspiration in the biological nervous system to predict its results. It is the appropriate strategy to model complex processes and it is able to learn from experience. Neural Networks generally require a big amount of data to be fully trained. Even though the experimental data was limiting, the results agree with the experimental values and the model was able to determine the importance of the bulge size for knockdown. The Fuzzy Logic Model is combining the strength of intuitive integration of prior knowledge with a sophisticated Global Genetic optimization Algorithm. After training the model, it was able to reproduce the experimental data, especially the correlation in a 3-dimensional space of the AU content score and 3' pairing score to the knockdown percentage.
- Adjustment of gene expression in specific tissues. - Tuning expression level accurately. - Predefine a construct to be used. These three paths were the inspiration behind creating miBEAT (miRNA Binding Site Engineering and Assembly Tool), to provide a strategy which makes possible to control the expression of genes in a specific way between tissues. To make this work, we tried to match the functionalities of the tool to our experimental project. Additionally, we provided a strategy that guides the user through the cloning process and allows them to use characterized standard parts sent to the MIT parts registry. Our complete work is present in the form of a graphical user interface called miBEAT. This tool combines and connects the output of the different models and scripts and then generates a suitable miTuner construct that will express the gene of interest, miGENE, up or down to the desired level. miBEAT consists of three subparts; miRockdown, miBS designer and mUTING. miRockdown is the subpart which contains two computational models that work on different concepts: Neural Network and Fuzzy Logic plus the experimentally obtained data. The models are sequentially associated with a script based on [http://www.targetscan.org/ Target Scan] algorithm. miRockdown takes as an input the desired knockdown percentage and the sequence of shRNAmir and gives out binding site parameters that are then compared with model predictions to finally generate the appropriate binding site. miBS designer is available as a stand alone for generating customized binding sites, but a modified version of it is also a part of miBEAT, in charge of generating more than 2000 different binding sites for every miRNA sequences, following more than 135 combinations of regions. mUTING provides the tissue specific targeting function to the GUI. It uses literature data for miRNA expression in various tissues and can output miRNA binding sites that could be used to differentiate between target and off target tissues. miRNA binding site featuresmiRNA are non-coding regulatory RNAs functioning as post-transcriptional gene silencers. After they are processed, they are usually 22 nucleotides long and they usually bind to the 3’UTR region of the mRNA (although they can also bind to the ORF or to the 5'UTR), forcing the mRNA into degradation or just repressing translation [Bartel, 2004]. In vegetal organisms, miRNA usually bind to the mRNA with extensive complementarity. In animals, interactions are more inexact, creating a lot of uncertainty in the in silico prediction of targets[?]. The seed of the miRNA is usually defined as the region centered in the nucleotides 2-7 in the 5’ end of the miRNA. For an efficient binding site extensive pairing is usually required between the seed and the corresponding part of the mRNA. The seed, and the corresponding pairing sequence of the mRNA are located inside the AGO protein. Common types of miRNA seeds: - 6mer (abundance 21.5%): only the nucleotides 2-7 of the miRNA match with the mRNA. - 7merA1 (abundance 15.1%): the nucleotides 2-7 match with the mRNA, and there is an adenine in position 1. - 7merm8 (abundance 25%): the nucleotides 2-8 match with the mRNA. - 8mer (abundance 19.8%): the nucleotides 2-8 match with the mRNA and there is an adenine in position 1. The percentages of abundance are calculated among conserved mammalian sites for a highly conserved miRNA (Friedman et al. 2008) 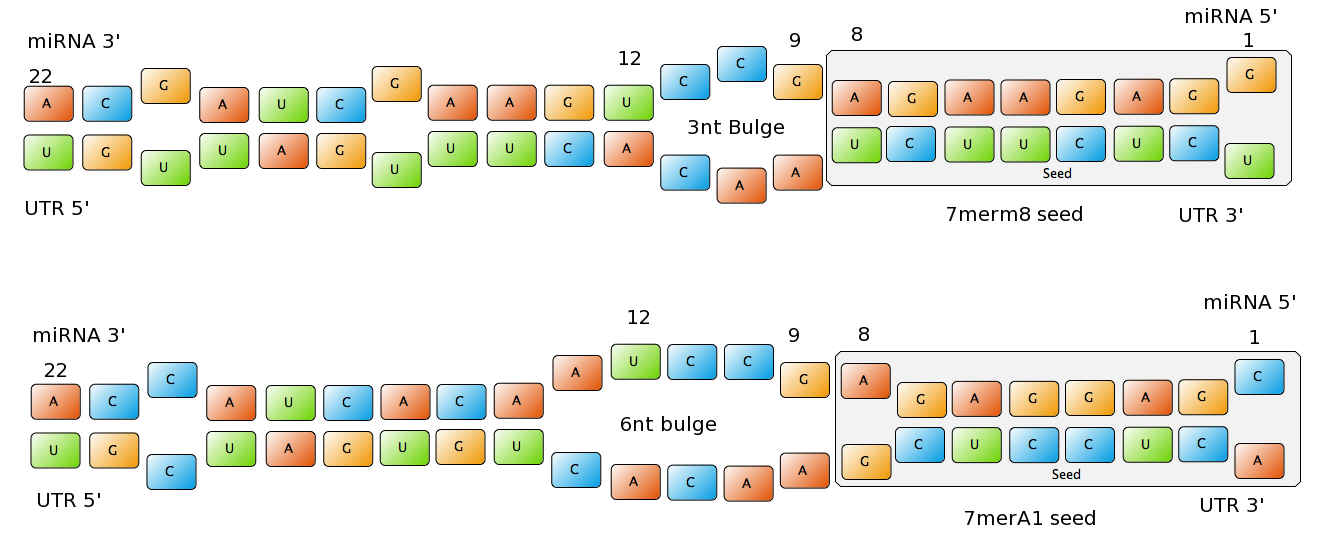 Figure 1: Interactions between two miRNAs and their binding sites. Notice the different types of seeds. Outside the seed, the existence of supplemental pairing (at least 3 contiguous nucleotides and at best centered in nucleotides 13-16 of the miRNA) stabilizes the bound complex and increases the efficacy of the binding site. Binding sites with a high local AU content around the binding site have proven to be more effective (possibly because of the destabilization of the mRNA secondary structure around the site). An arginine at position one of the binding site supposedly binds to a different protein of the RISC complex [Bartel, 2009], thus increasing the binding site efficiency significantly. Binding sites at the end or the beginning of the 3'UTR are more efficient. Binding sites within the first 15 nucleotides after the stop codon are not effective, since this region of the mRNA is inside the ribosome when translations stops. Thus a bound RISC complex in this region will dissociate after every round of translation and can not follow it's usual mode of action [Grimson et al., 2007].
Tissue specific miRNAsA useful supplement to achieving tuned gene expression in cells is the ability to specifically target tissues where this should be carried out. Tissue targeting has, for quite some time, been an important field of research that has drawn much attention and is central to gene therapy. miBEAT tool not only allows generation of binding sites that regulate the level of expression of a desired gene, but also employs strategies that help target the right tissue and exclude expression in others. This functionality is based on the principle of using tissue specific miRNA binding sites which can be introduced in the miTuner construct easily. A smart way to specifically target tissues is to exploit the presence and absence of tissue specific endogenous miRNA in the target to specifically express or exclude expression of the gene of interest in the target. We make use of two of strategies based on this principle, namely, on-targeting and off-targeting. The off-targeting concept has been applied previously [http://2010.igem.org/Team:Heidelberg/Modeling#References (Wenfang Shi et.al., 2008)] wherein an endogenous miRNA is selected such that it is not present in the target tissue (therefore the gene is expressed) and is present in all the off target tissues (knockdown of the transcripts). Thus the gene is specifically expressed in the target. In addition to the off-targeting strategy, we designed a new strategy, the on-targeting. In this case, the miRNA is present in the target tissue and excluded from the off-targets. The binding site for this miRNA is present within the 3'UTR of a repressor gene (in our case TET/O2) construct. The operator for the repressor in turn precedes the gene of interest (miGene) in the miTuner or miMeasure constructs. Therefore in the presence of miRNA in the cell, repressor is degraded and miGene is expressed while in off-targets repressor is translated and represses the expression of miGene.
|
 "
"