Team:EPF Lausanne/Project asaia
From 2010.igem.org
(→Antibiotic resistance) |
(→Asaia: A new chassis to fight mosquito-borne diseases) |
||
| (22 intermediate revisions not shown) | |||
| Line 23: | Line 23: | ||
<div CLASS="EPFL_content"> | <div CLASS="EPFL_content"> | ||
| - | |||
| - | |||
=Asaia: A new chassis to fight mosquito-borne diseases= | =Asaia: A new chassis to fight mosquito-borne diseases= | ||
| - | Asaia is a pink | + | [[Image:ID card.png|right|300px|caption]] |
| + | Asaia is a pink bacterium collected from tropical flowers in Thailand and Indonesia [ [[#References:|1]] ]. It has been proven to naturally live in the mosquito's intestinal tract and to have a high rate of transmission between mosquitos, both horizontally (from one mosquito to another) and vertically (from parent to offspring)[ [[#References:|2]] ]. The vertical and horizontal transmission as well as the cross-colonizing capacity of Asaia (as reported in literature [ [[#References:|3]] ]) could assure the spread of the transformed agent through a host population. It also appears to be quite specific to mosquitoes, and would therefore have little risk of spreading to other insects. However, there is experimental evidence that Asaia is able to cross-colonize different species of mosquitoes and does not exert any pathogenic effect on the host[ [[#References:|4]] ]. Therefore, Asaia is considered to be useful to the mosquito, because of its 100% prevalence as well as its preservation in laboratory conditions [ [[#References:|5]] ]. | ||
[[Image:Doctor.png|left|150px|caption]] | [[Image:Doctor.png|left|150px|caption]] | ||
| - | These properties make it a very good candidate for our project, which consists in blocking the cycle of [https://2010.igem.org/wiki/index.php?title=Team:EPF_Lausanne/Project_malaria Malaria] | + | These properties make it a very good candidate for our project, which consists in blocking the infection cycle of [https://2010.igem.org/wiki/index.php?title=Team:EPF_Lausanne/Project_malaria Malaria]. To assess if Asaia is actually a good organism to work on, our team at EPFL decided to study and characterize it: we worked on the strain SF2.1(Gfp) [ [[#References:|2]] ]which turned out is relatively easy to manipulate. Our team provides iGEM with a new chassis, and the necessary protocols for future teams that want to keep working on this promising organism. |
| - | We believe | + | We believe "Asaia" could be a good instrument in the fight against malaria and other mosquito-borne diseases. |
| - | + | ||
| + | In this page we summarize the main results related to growing and working with "Asaia". | ||
| - | + | == Do you want to work with Asaia?== | |
| + | No problem! You can find in the following tech-sheets all the information, protocols and tricks you need to start your own project. | ||
| - | + | *[https://static.igem.org/mediawiki/2010/3/3b/Growing_Asaia.pdf Growing Asaia] | |
| - | + | *[https://static.igem.org/mediawiki/2010/c/c5/Competent_cells.pdf Competent cells] | |
| - | + | *[https://static.igem.org/mediawiki/2010/a/a6/Transformation.pdf Transformation] | |
| - | + | *[https://static.igem.org/mediawiki/2010/9/95/Cloning_into_Asaia.pdf Cloning into Asaia] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
= Doubling Time = | = Doubling Time = | ||
| Line 62: | Line 55: | ||
= Antibiotic resistance = | = Antibiotic resistance = | ||
| - | We characterized Asaia's | + | We characterized Asaia's resistance against various antibiotics. |
| - | We had noticed previously that Asaia is naturally resistant to Ampicillin (Amp) and Chloroamphenicol (Cm) | + | We had noticed previously that Asaia is naturally resistant to Ampicillin (Amp) and Chloroamphenicol (Cm) using the standard concentrations generally used with ''E. coli''. |
| - | We tested the behaviour of Asaia to 7 | + | We tested the behaviour of Asaia to 7 other antibiotics like Tetracyclin (Tet) and Kanamycin (Kan) at various concentrations. |
| Line 70: | Line 63: | ||
| - | [[Image:Graph Resistance2.jpg|thumb|500px|Here you can see the doubling time | + | [[Image:Graph Resistance2.jpg|thumb|center|500px|Here you can see the doubling time as a function of antibiotic concentration. When Asaia didn't grow, the doubling time is arbitrarily set to 10h]] |
| - | [[Image:EPFL_ResArray.jpg|center|500px|Here is our plate with different | + | [[Image:EPFL_ResArray.jpg|thumb|center|500px||Here is our plate with different concentrations of antibiotics. Green holes are filled with growing Asaia.]] |
| + | |||
| + | =Microscope Images of Asaia= | ||
| + | We also had a look at Asaia (containing a plasmid expressing GFP) with the microscope. It looks very similar to <i>E.coli</i>. | ||
| + | [[Image:EPFL-Asaia-microscopy.jpg|thumb|center|500px||Brightfield (top) and a GFP fluorescence (bottom) picture of the Asaia strain containing the Plasmid pHM2-GFP]] | ||
= Compatibility with ''E. coli'' = | = Compatibility with ''E. coli'' = | ||
| - | We noticed that | + | [[Image:Copain.png|right|250px|caption]] |
| - | Therefore, we had to add Asaia's origin of replication to all our parts so they would function in both Asaia and ''E. coli'' | + | |
| + | We noticed that ''Asaia's'' origin of replication is compatible with ''E. coli'', but the opposite is not true. | ||
| + | Therefore, we had to add ''Asaia's'' origin of replication to all our parts so they would function in both ''Asaia'' and ''E. coli'' | ||
Latest revision as of 20:13, 27 October 2010


Contents |
Asaia: A new chassis to fight mosquito-borne diseases
Asaia is a pink bacterium collected from tropical flowers in Thailand and Indonesia [ 1 ]. It has been proven to naturally live in the mosquito's intestinal tract and to have a high rate of transmission between mosquitos, both horizontally (from one mosquito to another) and vertically (from parent to offspring)[ 2 ]. The vertical and horizontal transmission as well as the cross-colonizing capacity of Asaia (as reported in literature [ 3 ]) could assure the spread of the transformed agent through a host population. It also appears to be quite specific to mosquitoes, and would therefore have little risk of spreading to other insects. However, there is experimental evidence that Asaia is able to cross-colonize different species of mosquitoes and does not exert any pathogenic effect on the host[ 4 ]. Therefore, Asaia is considered to be useful to the mosquito, because of its 100% prevalence as well as its preservation in laboratory conditions [ 5 ].
These properties make it a very good candidate for our project, which consists in blocking the infection cycle of Malaria. To assess if Asaia is actually a good organism to work on, our team at EPFL decided to study and characterize it: we worked on the strain SF2.1(Gfp) [ 2 ]which turned out is relatively easy to manipulate. Our team provides iGEM with a new chassis, and the necessary protocols for future teams that want to keep working on this promising organism.
We believe "Asaia" could be a good instrument in the fight against malaria and other mosquito-borne diseases.
In this page we summarize the main results related to growing and working with "Asaia".
Do you want to work with Asaia?
No problem! You can find in the following tech-sheets all the information, protocols and tricks you need to start your own project.
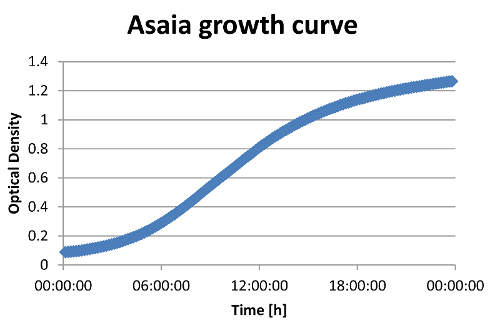
Doubling Time
We measured the growth curve of Asaia at different pH (pH2-pH7) and at 30°C and 37°C to figure out the optimal growth conditions. We found that the optimal Temperature is 30°C, and the optimal pH is 5. At this conditions we measured a doubling time of 2h40min.
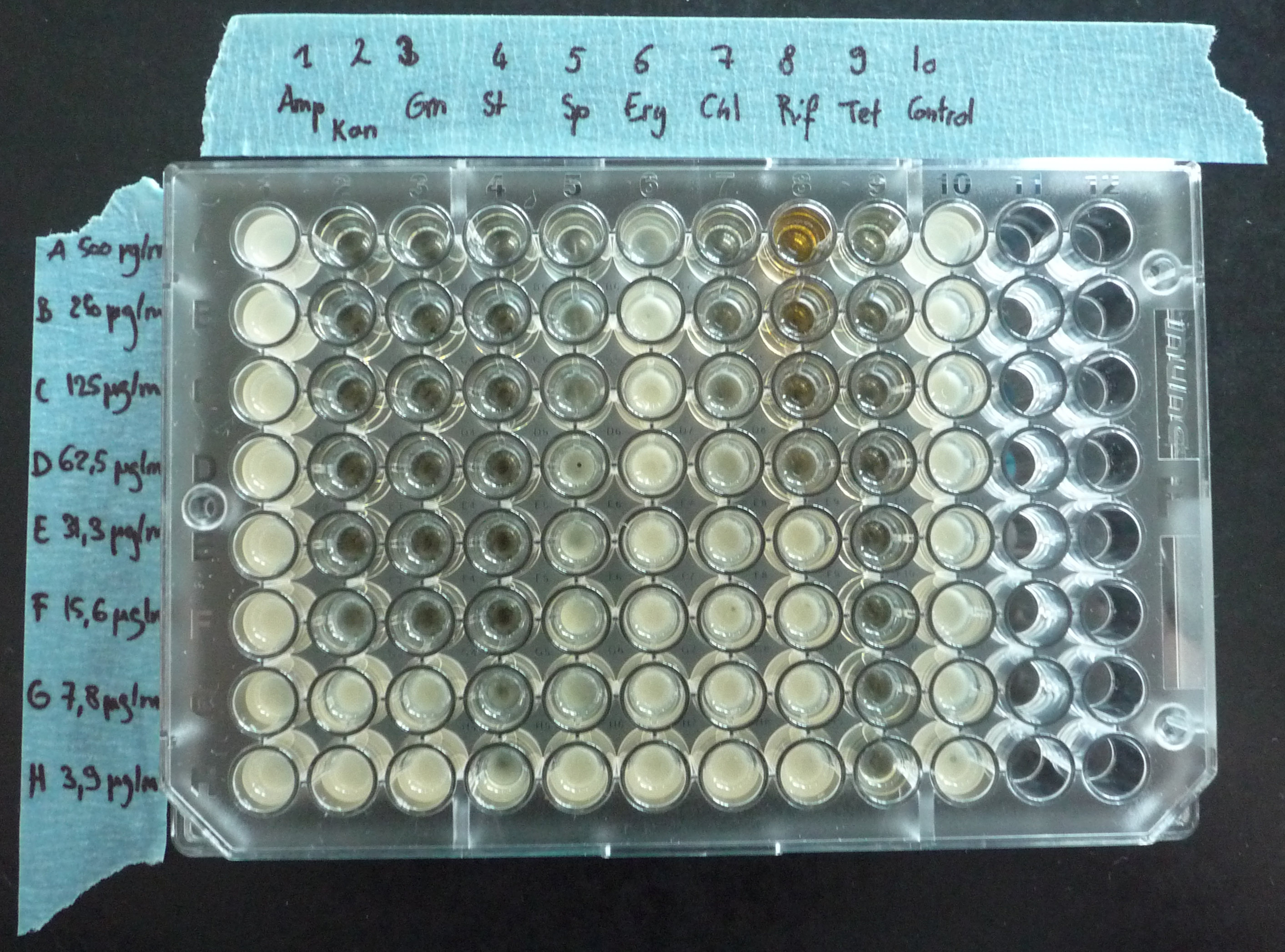
Antibiotic resistance
We characterized Asaia's resistance against various antibiotics. We had noticed previously that Asaia is naturally resistant to Ampicillin (Amp) and Chloroamphenicol (Cm) using the standard concentrations generally used with E. coli. We tested the behaviour of Asaia to 7 other antibiotics like Tetracyclin (Tet) and Kanamycin (Kan) at various concentrations.
Microscope Images of Asaia
We also had a look at Asaia (containing a plasmid expressing GFP) with the microscope. It looks very similar to E.coli.
Compatibility with E. coli
We noticed that Asaia's origin of replication is compatible with E. coli, but the opposite is not true. Therefore, we had to add Asaia's origin of replication to all our parts so they would function in both Asaia and E. coli
References
- [http://ijs.sgmjournals.org/cgi/reprint/51/2/559.pdf 1. Kazushige Katsura, Asaia siamensis sp. nov., an acetic acid bacterium in the α-Proteobacteria,International Journal of Systematic and Evolutionary Microbiology (2001)]
- [http://www.pnas.org/content/104/21/9047.long#sec-7. 2. Guido Favia, Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector ]
- [http://aem.asm.org/cgi/content/abstract/AEM.01336-10v1. 3. Crotti et al, Acetic acid bacteria, new emerging symbionts of insects (2010)]
- [http://aem.asm.org/cgi/reprint/AEM.01747-10v1?view=long&pmid=20851960. 4. Bessem et al, Typing of Asaia spp. bacterial symbionts in four mosquito species: molecular evidence for multiple infections (2010)]
- [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1885625/ 5. Guido et al,Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector (2007)]

 "
"