Team:Stockholm/Protocols
From 2010.igem.org
(→IgG protease activity assay) |
|||
| (41 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | ==Competent cells (Nina & Johan)== | ||
| + | ''From Morten Nørholm at the Department of Biochemistry & Biophysics Stockholm University'' | ||
| - | + | # Add 5 ml LB in two 50 ml falcon tubes. Add the top of a tip bacteria into the two 5 ml LB. Grow ON in shake incubator 37 degree C. | |
| + | # Subculture each 5 ml of starterculture into two 400 ml pre-warmed LB. Grow at 37 degree C until OD reaches 0.6. | ||
| + | # Put cells on ice for 20 min. | ||
| + | # Harvest cells at 4000 rpm for 20 min, 4 degree C. | ||
| + | # Discard supernatant if it looks clear (or spinn longer if it is cloudy). | ||
| + | # Resuspend pellet carefully in 500 50 mM CaCl2 for a 1000 ml cell culture (1/2 the orifinal volume). | ||
| + | # Put cells on ice 20 min. | ||
| + | # Repeat step 5. | ||
| + | # Resuspend cells in 16 ml CaCl2 + 15% Glycerol for a 800 ml starter culture (1:50 volume) | ||
| + | # Put metal blocks in -80 degree C. | ||
| + | # Snapfreeze 100 mikroL aliquots in ice-cold Epps (in pre-chilled blocks). Store in -80 degree C freezer. | ||
| - | + | ==Competent cells (Andreas & Mimmi)== | |
| - | 2. | + | ''Based and modified from the [http://openwetware.org/wiki/TOP10_chemically_competent_cells Top10 protocol by the Knight lab]'' |
| + | =====Materials===== | ||
| + | <blockquote>'''CCMB80 buffer''' | ||
| + | *10 mM KOAc pH 7.0 | ||
| + | *80 mM CaCl<sub>2</sub>.2H<sub>2</sub>O | ||
| + | *20 mM MnCl<sub>2</sub> | ||
| + | *10 mM MgCl<sub>2</sub>.6H<sub>2</sub>O | ||
| + | *10 % glycerol</blockquote> | ||
| + | =====Procedures===== | ||
| + | # Set an ON starter culture by inoculating 5 ml LB and incubating ON in 37 °C with 250 rpm rotary shaking. | ||
| + | # Inoculate 250 ml new LB with 2 ml from the ON culture and grow in 30 °C, 250 rpm until an OD<sub>600</sub> of ≈0.3. | ||
| + | #* Use a large, 1 l E-flask. | ||
| + | # Spin down cells at 3000 x ''g'' for 10 min in 4 °C. | ||
| + | # Remove supernatant and resuspend cells in 80 ml ice-cold '''CCMB80 buffer'''. Keep cells on ice for 20 min. | ||
| + | # Spin down cells again with same settings. Resuspend in 10 ml new CCMB80 buffer and keep on ice for 20 min. | ||
| + | # Aliquot 100 μl samples of competent cells into 1.5 ml vials and store in -80°C, or transform immediately. | ||
| - | + | ==Transformation (Nina & Johan)== | |
| + | ''From NEB 5-alpha competent E.coli (High Efficiency) NEW ENGLAND BioLabs'' | ||
| - | + | # Thaw a tube of NEB 5-alpha competent E.coli cells on ice for 10 min. | |
| + | # Add 1-5 μl containing 1 pg-100 ng of plasmid DNA to the cell mixture. Carefully flick the tube 4-5 times to mix the cells and DNA. | ||
| + | # Place the mixture on ice for 30 min. | ||
| + | # Heat shock at 42°C for 30 sec. | ||
| + | # Place on ice for 5 min. | ||
| + | # Pipette 950 μl of room temp. SOC into the mixture. | ||
| + | # Place at 37°C for 30-60 min, 250 rpm. | ||
| + | # Warm selection plates to 37°C. | ||
| + | # Perform 10-fold serial dilutions in SOC (if necessary) | ||
| + | # Spread 50-100 μl of each sample onto a selection plate and incubate overnight at 37°C. | ||
| - | + | ==Transformation (Andreas & Mimmi)== | |
| + | # Add 1 μl plasmid to 100 μl thawed, competent cells of choice. Hold cells on ice for 30 min. | ||
| + | # Heat-shock cells for 55 sec in 42 °C. Return to ice. | ||
| + | # Add 900 μl LB medium and grow cells in 37°C, 250 rpm for 1 hour. | ||
| + | #* This allows cells to start expressing antibiotic resistance gene(s). | ||
| + | # Spin down cells at full speed (≈13.000 x ''g'') for 15 sec. | ||
| + | # Remove 900 μl from the supernatant and gently resuspend pellet in remaining 100 μl. | ||
| + | # Plate 100 μl cells onto an LB agar plate with appropriate antibiotic(s). | ||
| + | # Incubate in 37 °C ON. | ||
| - | + | ==Quick transformation (Andreas & Mimmi)== | |
| + | # Add 1-3 μl plasmid to 100 μl thawed, competent Top10 cells. Hold cells on ice for 5 min. | ||
| + | # Heat-shock cells for 30 sec in 42 °C. Return to ice. | ||
| + | # Plate cells onto a pre-heated (37 °C) LB agar plate with appropriate antibiotic(s). | ||
| + | # Incubate in 37 °C ON. | ||
| - | + | ==Colony PCR verification (Andreas & Mimmi)== | |
| + | # Pick four colonies and resuspend each in 10 μl LB. | ||
| + | #* Let incubate in RT while preparing PCR tubes. | ||
| + | # Prepare each illustra PuReTaq Ready-To-Go PCR (GE Healthcare) tube as follows: | ||
| + | #* 1.0 μl 10 μM forward primer | ||
| + | #* 1.0 μl 10 μM reverse primer | ||
| + | #* 22.5 μl dH<sub>2</sub>O | ||
| + | #* 0.5 μl cell suspension (template DNA) to a '''final volume of 25 μl'''. Vortex to mix. | ||
| + | # Run PCR amplification. | ||
| + | #* Denaturation: 95 °C - 10 min | ||
| + | #* 30 cycles | ||
| + | ## Denaturation: 95 °C - 30 s | ||
| + | ## Annealing: 55 °C - 30 s | ||
| + | ## Elongation: 72 °C - ''Calculate from expected sequence length, '''≈1 min/kb''''' | ||
| + | #* Elongation: 72 °C - 10 min | ||
| + | # Analyze PCR products by agarose gel electrophoresis. | ||
| - | + | ==Site-directed mutagenesis== | |
| + | ''Based on the QuikChange® Site-Directed Mutagenesis Kit'' | ||
| + | * Design two complimentary oligonucleotides containing the desired mutation with https://www.genomics.agilent.com/CollectionSubpage.aspx?PageType=Tool&SubPageType=ToolQCPD&PageID=15 | ||
| + | # Prepare PCR (amounts for 100 µl reaction) | ||
| + | #* 10 ng dsDNA template | ||
| + | #* 125 ng primer #1 | ||
| + | #* 125 ng primer #2 | ||
| + | #* 84 µl ddH2O | ||
| + | #* 2 µl 10 mM dNTPs | ||
| + | #* 10 µl 10x Pfu reaction buffer | ||
| + | #* 2 µl Pfu Turbo DNA polymerase | ||
| + | # Run PCR | ||
| + | ## 95 °C - 30 sec | ||
| + | ## 22 cycles of | ||
| + | ### 95 °C - 30 sec | ||
| + | ### 55 °C - 1 min | ||
| + | ### 68 °C - 1 min/kb plasmid | ||
| + | ## 4 °C - ∞ | ||
| + | # Make sure the reaction is ≤ 37 °C before proceeding | ||
| + | # Add 2 µl (for 100 µl reaction) of Dpn I restriction enzyme and mix thoroughly | ||
| + | # Spin down for 1 min in microcentrifuge | ||
| + | # Immediately incubate at 37 °C for 1 hour or more (more gives less background) | ||
| - | + | ---- | |
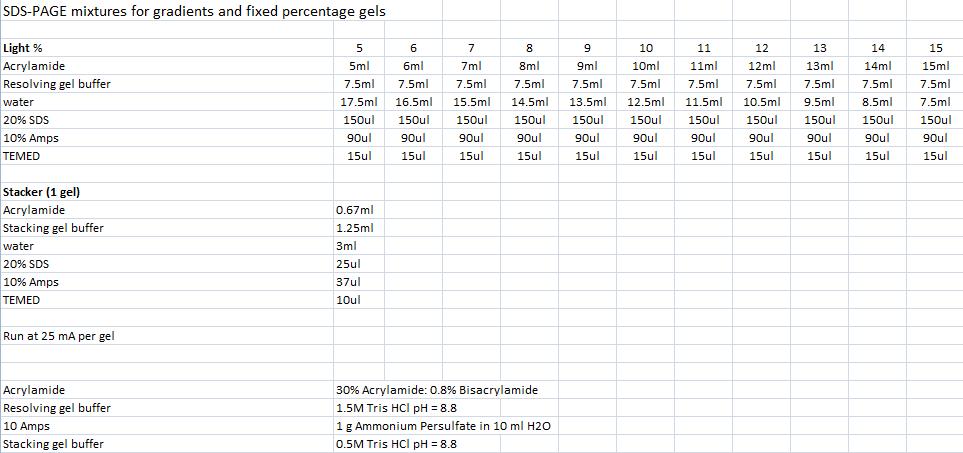
| + | ==SDS-PAGE mixtures (Nina & Johan)== | ||
| + | ''From Robert Daniels at the Department of Biochemistry & Biophysics Stockholm University'' | ||
| - | + | [[Image:SDS.jpg]] | |
| - | + | ---- | |
| + | ==Mini prep(Nina & Johan)== | ||
| + | ''Based on the QIAprep Spin Miniprep Kit Using a Microcentrifuge'' | ||
| + | 1. Centrifuge sample in 4 °C, 10 min, 4000 rpm and resuspend pelleted bacterial cells with Buffer P1 (250 ul/5ml bacterial sample) and transfer to a microcentrifuge tube. | ||
| - | == | + | 2. Add 250 ul buffer P2, invert the tubes 4-6 times. |
| - | # | + | |
| - | # | + | 3. Add 350 ul buffer N3, invert the tubes 4-6 times with powerful strokes. |
| - | # | + | |
| - | #* | + | 4. Centrifuge 10 min, 13000 rpm in a table-top microcentrifuge. |
| - | # Spin down | + | |
| - | # | + | 5. Transfer the supernatant to the QIAprep spin column. |
| - | # | + | |
| + | 6. Centrifuge for 1 min, 13000 rpm. Discard the flow-through. | ||
| + | |||
| + | 7. Add 0.5 ml buffer PB and centrifuge for 1 min, 13000 rpm. Discard the flow-through. | ||
| + | |||
| + | 8. Add 0.75 ml buffer PE and centrifuge for 1 min, 13000 rpm. | ||
| + | |||
| + | 9. Discard the flow-through and centrifuge again the same way. | ||
| + | |||
| + | 10. Place the column in a clean 1.5 ml microcentrifuge tube. Add 35 ul water to the center of the QIAprep spin column, let stand for 1 min, and centrifuge for 1 min, 13000 rpm. | ||
| + | |||
| + | ---- | ||
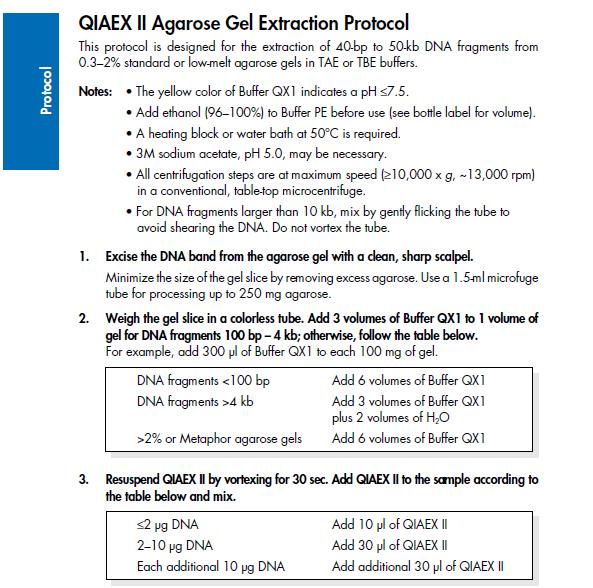
| + | ==Agarose gel clean up (Nina & Johan)== | ||
| + | |||
| + | ''Based on the QIAprep II Handbook'' | ||
| + | |||
| + | [[Image:Protocol.jpg]] | ||
| + | |||
| + | [[Image:Protocol1.jpg]] | ||
| + | |||
| + | [[Image:Protocol2.jpg]] | ||
| + | |||
| + | [[Image:Protocol3.jpg]] | ||
| + | |||
| + | ==IgG protease activity assay== | ||
| + | |||
| + | Based on an experiment by [http://www.ncbi.nlm.nih.gov/pubmed/20441890 Abuknesha, ''et al'' (2010)]. | ||
| + | |||
| + | ===Materials=== | ||
| + | |||
| + | *10 ml culture tubes | ||
| + | *200 ml E-flasks | ||
| + | *10 ml Falcon tubes | ||
| + | *IPTG | ||
| + | *Protease dilution buffer | ||
| + | *Eppendorf tubes | ||
| + | *[http://en.wikipedia.org/wiki/McFarland_standards 0.5 McFarland standard] | ||
| + | **0.05 ml 1 % BaCl<sub>2</sub> | ||
| + | **9.95 ml 1 %/0.18 M H<sub>2</sub>SO<sub>4</sub> | ||
| + | *[http://openwetware.org/wiki/PBS Phosphate buffered saline (PBS), pH 7.4] | ||
| + | *Mouse IgG-Agarose (1 mg IgG/ml) (Sigma-Aldrich) | ||
| + | *Anti-mouse IgG peroxidase conjugate (Sigma Aldrich) | ||
| + | *SureBlue™ TMB Microwell Peroxidase Substrate | ||
| + | |||
| + | ===Procedures=== | ||
| + | |||
| + | ====Day 0==== | ||
| + | #Set ON cultures in 5 ml LB + 100 μg/ml Amp, 37 °C, 225 rpm | ||
| + | #*BL21 IgGp | ||
| + | #*BL21 SOD (or other negative control) | ||
| + | |||
| + | ====Day 1==== | ||
| + | #Inoculate 200 μl of each ON culture (x2 for IgGp) into 20 ml fresh LB with 100 μg/ml Amp. Grow at 37 °C, 225 rpm until an OD<sub>600</sub> of ≈0.5. | ||
| + | #Induce protein expression by adding IPTG to a final concentration of 0.3 mM (e.g. 60 μl 0.1 M IPTG) to one of each culture type. Leave the second IgGp culture uninduced (negative control). Continue incubation for two hours. | ||
| + | #Transfer cultures to 10 ml Falcon tubes and spin down cells at 4,400 x g for 10 min. | ||
| + | #Resuspend enough pellet in 5 ml sterile PBS to yield a cell density of 1.5 x 10<sup>8</sup> CFU ml<sup>-1</sup> according to the McFarland standard 0.5. Make a 1000x dilution of each culture with PBS to 1.5 x 10<sup>5</sup>. | ||
| + | #Sonicate cells and spin down cell debris at 4,400 x g for 10 min. Recover and save supernatants for protease assay. | ||
| + | #*Uninduced IgGp | ||
| + | #**1.5 x 108 CFU ml-1 | ||
| + | #**1.5 x 105 CFU ml-1 | ||
| + | #*Induced IgGp | ||
| + | #**1.5 x 108 CFU ml-1 | ||
| + | #**1.5 x 105 CFU ml-1 | ||
| + | #*Induced SOD | ||
| + | #**1.5 x 108 CFU ml-1 | ||
| + | #**1.5 x 105 CFU ml-1 | ||
| + | #Prepare IgG peroxidase dilutions by diluting the stock solution in PBS to 1:500 and 1:1000. | ||
| + | #Apply 30 μl IgG-Agarose suspension to 14 Eppendorf tubes, two for each protein extract dilution and two negative controls. | ||
| + | #Bind secondary antibodies by adding 100 μl anti-mouse IgG peroxidase, each dilution (1:500, 1:1000) to 7 tubes. Allow antibody binding by incubating at room temperature with tilt mixing for 30 min. | ||
| + | #Spin down IgG-Agarose at 13,000 x g for 3 min and wash 3 times with 200 μl PBS. Spin down again and remove supernatant. | ||
| + | #Add 100 μl of the protease extracts to the corresponding tubes (12 tubes). Add 100 μl PBS to the remaining two negative control tubes. Incubate at 37 °C ON (≈16 h) with tilt mixing. | ||
| + | |||
| + | ====Day 2==== | ||
| + | #Spin down the IgG-Agarose at 13,000 x g, 3 min. Transfer 80 μl of each supernatant (no pellet!) to new Eppendorf tubes. | ||
| + | #Add 100 μl Sure Blue™ peroxidase substrate solution. Incubate in room temperature and tap gently until a blue color develops (≈30 min). | ||
| + | #Stop reaction by adding 100 μl 1 M HCl. | ||
| + | #Read color intensity/absorbance at 450 nm. | ||
Latest revision as of 09:20, 22 October 2010
Competent cells (Nina & Johan)
From Morten Nørholm at the Department of Biochemistry & Biophysics Stockholm University
- Add 5 ml LB in two 50 ml falcon tubes. Add the top of a tip bacteria into the two 5 ml LB. Grow ON in shake incubator 37 degree C.
- Subculture each 5 ml of starterculture into two 400 ml pre-warmed LB. Grow at 37 degree C until OD reaches 0.6.
- Put cells on ice for 20 min.
- Harvest cells at 4000 rpm for 20 min, 4 degree C.
- Discard supernatant if it looks clear (or spinn longer if it is cloudy).
- Resuspend pellet carefully in 500 50 mM CaCl2 for a 1000 ml cell culture (1/2 the orifinal volume).
- Put cells on ice 20 min.
- Repeat step 5.
- Resuspend cells in 16 ml CaCl2 + 15% Glycerol for a 800 ml starter culture (1:50 volume)
- Put metal blocks in -80 degree C.
- Snapfreeze 100 mikroL aliquots in ice-cold Epps (in pre-chilled blocks). Store in -80 degree C freezer.
Competent cells (Andreas & Mimmi)
Based and modified from the [http://openwetware.org/wiki/TOP10_chemically_competent_cells Top10 protocol by the Knight lab]
Materials
CCMB80 buffer
- 10 mM KOAc pH 7.0
- 80 mM CaCl2.2H2O
- 20 mM MnCl2
- 10 mM MgCl2.6H2O
- 10 % glycerol
Procedures
- Set an ON starter culture by inoculating 5 ml LB and incubating ON in 37 °C with 250 rpm rotary shaking.
- Inoculate 250 ml new LB with 2 ml from the ON culture and grow in 30 °C, 250 rpm until an OD600 of ≈0.3.
- Use a large, 1 l E-flask.
- Spin down cells at 3000 x g for 10 min in 4 °C.
- Remove supernatant and resuspend cells in 80 ml ice-cold CCMB80 buffer. Keep cells on ice for 20 min.
- Spin down cells again with same settings. Resuspend in 10 ml new CCMB80 buffer and keep on ice for 20 min.
- Aliquot 100 μl samples of competent cells into 1.5 ml vials and store in -80°C, or transform immediately.
Transformation (Nina & Johan)
From NEB 5-alpha competent E.coli (High Efficiency) NEW ENGLAND BioLabs
- Thaw a tube of NEB 5-alpha competent E.coli cells on ice for 10 min.
- Add 1-5 μl containing 1 pg-100 ng of plasmid DNA to the cell mixture. Carefully flick the tube 4-5 times to mix the cells and DNA.
- Place the mixture on ice for 30 min.
- Heat shock at 42°C for 30 sec.
- Place on ice for 5 min.
- Pipette 950 μl of room temp. SOC into the mixture.
- Place at 37°C for 30-60 min, 250 rpm.
- Warm selection plates to 37°C.
- Perform 10-fold serial dilutions in SOC (if necessary)
- Spread 50-100 μl of each sample onto a selection plate and incubate overnight at 37°C.
Transformation (Andreas & Mimmi)
- Add 1 μl plasmid to 100 μl thawed, competent cells of choice. Hold cells on ice for 30 min.
- Heat-shock cells for 55 sec in 42 °C. Return to ice.
- Add 900 μl LB medium and grow cells in 37°C, 250 rpm for 1 hour.
- This allows cells to start expressing antibiotic resistance gene(s).
- Spin down cells at full speed (≈13.000 x g) for 15 sec.
- Remove 900 μl from the supernatant and gently resuspend pellet in remaining 100 μl.
- Plate 100 μl cells onto an LB agar plate with appropriate antibiotic(s).
- Incubate in 37 °C ON.
Quick transformation (Andreas & Mimmi)
- Add 1-3 μl plasmid to 100 μl thawed, competent Top10 cells. Hold cells on ice for 5 min.
- Heat-shock cells for 30 sec in 42 °C. Return to ice.
- Plate cells onto a pre-heated (37 °C) LB agar plate with appropriate antibiotic(s).
- Incubate in 37 °C ON.
Colony PCR verification (Andreas & Mimmi)
- Pick four colonies and resuspend each in 10 μl LB.
- Let incubate in RT while preparing PCR tubes.
- Prepare each illustra PuReTaq Ready-To-Go PCR (GE Healthcare) tube as follows:
- 1.0 μl 10 μM forward primer
- 1.0 μl 10 μM reverse primer
- 22.5 μl dH2O
- 0.5 μl cell suspension (template DNA) to a final volume of 25 μl. Vortex to mix.
- Run PCR amplification.
- Denaturation: 95 °C - 10 min
- 30 cycles
- Denaturation: 95 °C - 30 s
- Annealing: 55 °C - 30 s
- Elongation: 72 °C - Calculate from expected sequence length, ≈1 min/kb
- Elongation: 72 °C - 10 min
- Analyze PCR products by agarose gel electrophoresis.
Site-directed mutagenesis
Based on the QuikChange® Site-Directed Mutagenesis Kit
- Design two complimentary oligonucleotides containing the desired mutation with https://www.genomics.agilent.com/CollectionSubpage.aspx?PageType=Tool&SubPageType=ToolQCPD&PageID=15
- Prepare PCR (amounts for 100 µl reaction)
- 10 ng dsDNA template
- 125 ng primer #1
- 125 ng primer #2
- 84 µl ddH2O
- 2 µl 10 mM dNTPs
- 10 µl 10x Pfu reaction buffer
- 2 µl Pfu Turbo DNA polymerase
- Run PCR
- 95 °C - 30 sec
- 22 cycles of
- 95 °C - 30 sec
- 55 °C - 1 min
- 68 °C - 1 min/kb plasmid
- 4 °C - ∞
- Make sure the reaction is ≤ 37 °C before proceeding
- Add 2 µl (for 100 µl reaction) of Dpn I restriction enzyme and mix thoroughly
- Spin down for 1 min in microcentrifuge
- Immediately incubate at 37 °C for 1 hour or more (more gives less background)
SDS-PAGE mixtures (Nina & Johan)
From Robert Daniels at the Department of Biochemistry & Biophysics Stockholm University
Mini prep(Nina & Johan)
Based on the QIAprep Spin Miniprep Kit Using a Microcentrifuge
1. Centrifuge sample in 4 °C, 10 min, 4000 rpm and resuspend pelleted bacterial cells with Buffer P1 (250 ul/5ml bacterial sample) and transfer to a microcentrifuge tube.
2. Add 250 ul buffer P2, invert the tubes 4-6 times.
3. Add 350 ul buffer N3, invert the tubes 4-6 times with powerful strokes.
4. Centrifuge 10 min, 13000 rpm in a table-top microcentrifuge.
5. Transfer the supernatant to the QIAprep spin column.
6. Centrifuge for 1 min, 13000 rpm. Discard the flow-through.
7. Add 0.5 ml buffer PB and centrifuge for 1 min, 13000 rpm. Discard the flow-through.
8. Add 0.75 ml buffer PE and centrifuge for 1 min, 13000 rpm.
9. Discard the flow-through and centrifuge again the same way.
10. Place the column in a clean 1.5 ml microcentrifuge tube. Add 35 ul water to the center of the QIAprep spin column, let stand for 1 min, and centrifuge for 1 min, 13000 rpm.
Agarose gel clean up (Nina & Johan)
Based on the QIAprep II Handbook
IgG protease activity assay
Based on an experiment by [http://www.ncbi.nlm.nih.gov/pubmed/20441890 Abuknesha, et al (2010)].
Materials
- 10 ml culture tubes
- 200 ml E-flasks
- 10 ml Falcon tubes
- IPTG
- Protease dilution buffer
- Eppendorf tubes
- [http://en.wikipedia.org/wiki/McFarland_standards 0.5 McFarland standard]
- 0.05 ml 1 % BaCl2
- 9.95 ml 1 %/0.18 M H2SO4
- [http://openwetware.org/wiki/PBS Phosphate buffered saline (PBS), pH 7.4]
- Mouse IgG-Agarose (1 mg IgG/ml) (Sigma-Aldrich)
- Anti-mouse IgG peroxidase conjugate (Sigma Aldrich)
- SureBlue™ TMB Microwell Peroxidase Substrate
Procedures
Day 0
- Set ON cultures in 5 ml LB + 100 μg/ml Amp, 37 °C, 225 rpm
- BL21 IgGp
- BL21 SOD (or other negative control)
Day 1
- Inoculate 200 μl of each ON culture (x2 for IgGp) into 20 ml fresh LB with 100 μg/ml Amp. Grow at 37 °C, 225 rpm until an OD600 of ≈0.5.
- Induce protein expression by adding IPTG to a final concentration of 0.3 mM (e.g. 60 μl 0.1 M IPTG) to one of each culture type. Leave the second IgGp culture uninduced (negative control). Continue incubation for two hours.
- Transfer cultures to 10 ml Falcon tubes and spin down cells at 4,400 x g for 10 min.
- Resuspend enough pellet in 5 ml sterile PBS to yield a cell density of 1.5 x 108 CFU ml-1 according to the McFarland standard 0.5. Make a 1000x dilution of each culture with PBS to 1.5 x 105.
- Sonicate cells and spin down cell debris at 4,400 x g for 10 min. Recover and save supernatants for protease assay.
- Uninduced IgGp
- 1.5 x 108 CFU ml-1
- 1.5 x 105 CFU ml-1
- Induced IgGp
- 1.5 x 108 CFU ml-1
- 1.5 x 105 CFU ml-1
- Induced SOD
- 1.5 x 108 CFU ml-1
- 1.5 x 105 CFU ml-1
- Uninduced IgGp
- Prepare IgG peroxidase dilutions by diluting the stock solution in PBS to 1:500 and 1:1000.
- Apply 30 μl IgG-Agarose suspension to 14 Eppendorf tubes, two for each protein extract dilution and two negative controls.
- Bind secondary antibodies by adding 100 μl anti-mouse IgG peroxidase, each dilution (1:500, 1:1000) to 7 tubes. Allow antibody binding by incubating at room temperature with tilt mixing for 30 min.
- Spin down IgG-Agarose at 13,000 x g for 3 min and wash 3 times with 200 μl PBS. Spin down again and remove supernatant.
- Add 100 μl of the protease extracts to the corresponding tubes (12 tubes). Add 100 μl PBS to the remaining two negative control tubes. Incubate at 37 °C ON (≈16 h) with tilt mixing.
Day 2
- Spin down the IgG-Agarose at 13,000 x g, 3 min. Transfer 80 μl of each supernatant (no pellet!) to new Eppendorf tubes.
- Add 100 μl Sure Blue™ peroxidase substrate solution. Incubate in room temperature and tap gently until a blue color develops (≈30 min).
- Stop reaction by adding 100 μl 1 M HCl.
- Read color intensity/absorbance at 450 nm.
 "
"