Team:UNIPV-Pavia/Project/IntYeast
From 2010.igem.org
| (One intermediate revision not shown) | |||
| Line 38: | Line 38: | ||
<p align="center" valign="top"><br> | <p align="center" valign="top"><br> | ||
| - | <font size=" | + | <font size="5" align="center" valign="top"><b>Implementation and Results: Integrative standard vector for yeast</b></font> |
</p> | </p> | ||
<br> | <br> | ||
| Line 89: | Line 89: | ||
#<partinfo>BBa_K300980</partinfo>, provided by Mr Gene DNA synthesis service (www.mrgene.com), was excised from its original shipping vector (pMA) through digestion with MfeI (Fermentas) and NsiI (Fermentas) restriction enzymes. It was then isolated through a 1% agarose gel electrophoresis and gel-extracted (Macherey-Nagel NucleoSpin Extract II). | #<partinfo>BBa_K300980</partinfo>, provided by Mr Gene DNA synthesis service (www.mrgene.com), was excised from its original shipping vector (pMA) through digestion with MfeI (Fermentas) and NsiI (Fermentas) restriction enzymes. It was then isolated through a 1% agarose gel electrophoresis and gel-extracted (Macherey-Nagel NucleoSpin Extract II). | ||
| - | #<partinfo>BBa_I763007</partinfo>, available in the Registry, was digested with EcoRI-PstI (Roche), | + | #<partinfo>BBa_I763007</partinfo>, available in the Registry, was digested with EcoRI-PstI (Roche), run on agarose gel and its <partinfo>pSB1A2</partinfo> vector was gel-extracted as described above. |
#Digested <partinfo>BBa_K300980</partinfo> and <partinfo>pSB1A2</partinfo> have compatible ends (EcoRI-MfeI and PstI-NsiI). They were ligated with T4 Ligase (Roche) and transformed into competent TOP10 E. coli strain (<partinfo>BBa_V1009</partinfo>) which were then plated on Ampicillin (100 ug/ml) plates. | #Digested <partinfo>BBa_K300980</partinfo> and <partinfo>pSB1A2</partinfo> have compatible ends (EcoRI-MfeI and PstI-NsiI). They were ligated with T4 Ligase (Roche) and transformed into competent TOP10 E. coli strain (<partinfo>BBa_V1009</partinfo>) which were then plated on Ampicillin (100 ug/ml) plates. | ||
#Positive transformants were identified by colony PCR with VF2 (<partinfo>BBa_G00100</partinfo>) and VR (<partinfo>BBa_G00101</partinfo>) standard primers and by restriction mapping with EcoRI (Roche) or NsiI (Fermentas). The yielded plasmid had <partinfo>BBa_B0033</partinfo> flanked by EcoRI and SpeI. | #Positive transformants were identified by colony PCR with VF2 (<partinfo>BBa_G00100</partinfo>) and VR (<partinfo>BBa_G00101</partinfo>) standard primers and by restriction mapping with EcoRI (Roche) or NsiI (Fermentas). The yielded plasmid had <partinfo>BBa_B0033</partinfo> flanked by EcoRI and SpeI. | ||
Latest revision as of 15:28, 26 October 2010
|
Integrative standard vector for yeastMaterials and MethodsStrain: the S. cerevisiae S288C strain (<partinfo>BBa_K300979</partinfo>, genotype: MATα ρ° trp1-0) was purchased from Open Biosystems. Construction of BBa_K300001:
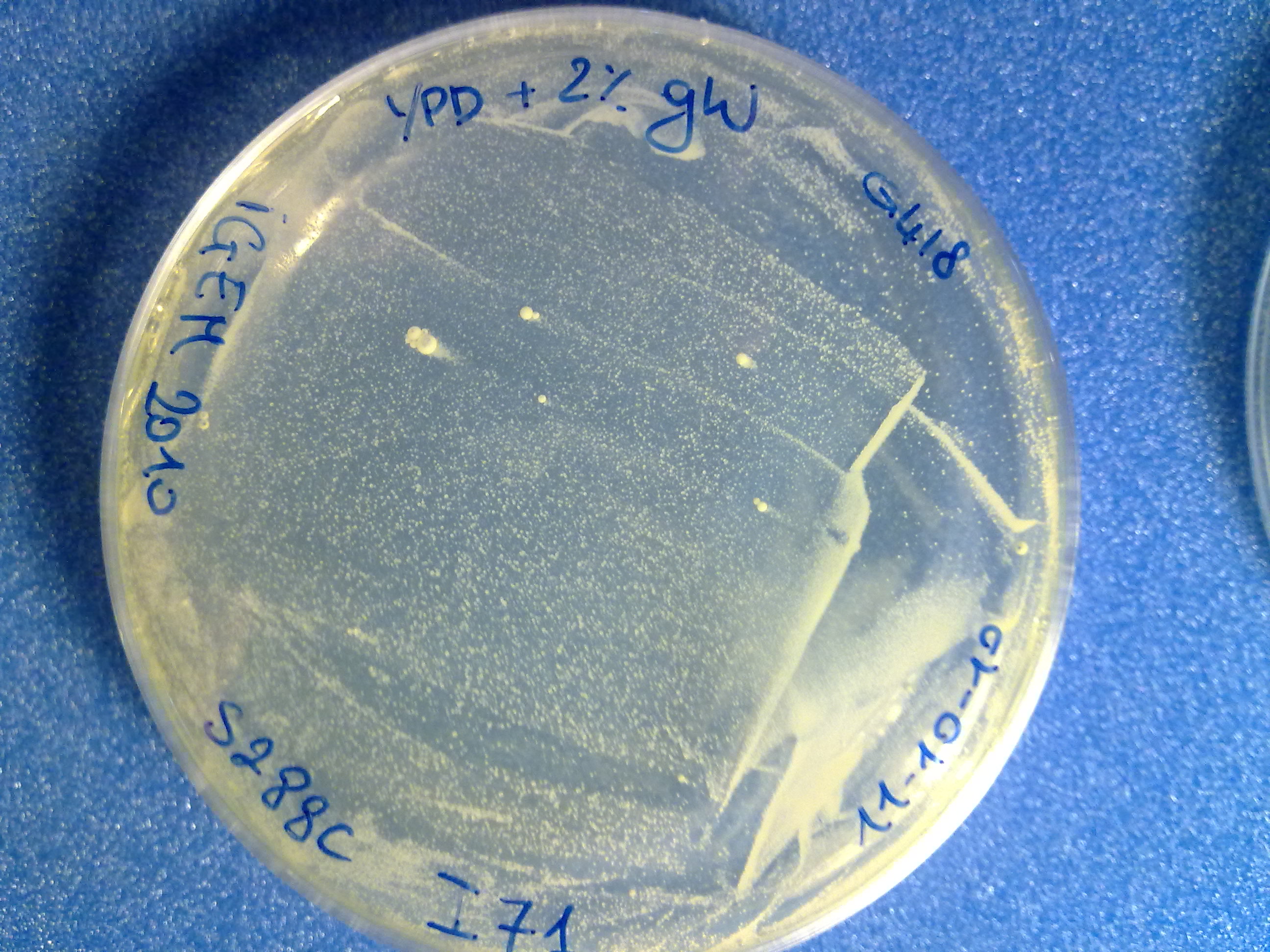
[1] http://openwetware.org/wiki/High_Efficiency_Transformation [2] Guldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1996), A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Research, Vol. 24, No. 13 2519–2524. ResultsThe transformed inserts and their integration efficiency in S288C are listed here:
The colony count was quite difficult (see pictures), but it is useful to have an estimation of the integration efficiency. DiscussionThe obtained results suggest that the integrative vector actually works and that the selection marker is highly specific (no colonies appeared on the "no DNA" plate). Although this integrative vector has already given promising integration results, a lot of work still remains to do:
As reported for the integrative vector for E. coli, the main information about the usage of this integrative vector for yeast is shared in the Registry. We hope to have positively contributed to the diffusion of standard biological parts for yeast, a very interesting chassis for synthetic biology for which only a limited number of parts are available in the Registry and only a few of them has been exaustively characterized.
|
 "
"