Team:Michigan/Oil Sands August September
From 2010.igem.org
(→9/30/2010) |
(→9/30/2010) |
||
| (22 intermediate revisions not shown) | |||
| Line 94: | Line 94: | ||
| - | | - | ||
|} | |} | ||
| + | |||
| + | [[Team:Michigan/Oil_Sands | Back to Oil Sands Main Notebook]] | ||
==8/1/2010== | ==8/1/2010== | ||
| Line 99: | Line 101: | ||
'''Nanodrop of pBAD and GFP verification''' | '''Nanodrop of pBAD and GFP verification''' | ||
| - | This miniprep was performed for the two cultures started last night according the the MODIFIED miniprep protocol. A frozen stock was made of the pBAD biobrick and placed in the - | + | This miniprep was performed for the two cultures started last night according the the MODIFIED miniprep protocol. A frozen stock was made of the pBAD biobrick and placed in the -80°C freezer. |
The DNA concentrations reported by the nanodrop are as follows: | The DNA concentrations reported by the nanodrop are as follows: | ||
| Line 123: | Line 125: | ||
[[Image:8-2-2010_gel_of_pbad_and_gfp_annotated.jpg|400px]] | [[Image:8-2-2010_gel_of_pbad_and_gfp_annotated.jpg|400px]] | ||
| - | *Lane 1: | + | *Lane 1: Invitrogen 1 kb Plus ladder |
| - | *Lane 2: GFP #3 cut with XbaI and PstI (GFP: 720 bp Backbone: 2079 bp) | + | *Lane 2: GFP #3 cut with XbaI and PstI (GFP: 720 bp; Backbone: 2079 bp) |
*Lane 3: GFP #4 cut with EcoRI and XbaI | *Lane 3: GFP #4 cut with EcoRI and XbaI | ||
*Lane 4: uncut GFP plamid (only 1 uL left so did not show up on gel) | *Lane 4: uncut GFP plamid (only 1 uL left so did not show up on gel) | ||
| - | *Lane 5: pBAD cut with EcoRI and SpeI(pBAD: 1210 bp | + | *Lane 5: pBAD cut with EcoRI and SpeI (pBAD: 1210 bp; Backbone: 4425 bp) |
*Lane 6: pBAD cut with SpeI and PstI | *Lane 6: pBAD cut with SpeI and PstI | ||
*Lane 7: uncut pBAD (loaded very small amount into gel by accident) | *Lane 7: uncut pBAD (loaded very small amount into gel by accident) | ||
| Line 143: | Line 145: | ||
==8/3/2010== | ==8/3/2010== | ||
| - | '''Minimal Media | + | '''Minimal Media''' |
Minimal media was mixed today according to the following [[Media:8-3-2010_media_recipies_updated.pdf|recipes]] (also found in the media recipe section of the notebooks): | Minimal media was mixed today according to the following [[Media:8-3-2010_media_recipies_updated.pdf|recipes]] (also found in the media recipe section of the notebooks): | ||
| - | '''Culturing Bacteria in Naphthenic Acids ( | + | '''Culturing Bacteria in Naphthenic Acids (NAs)''' |
The following 9 cultures were started to test bacteria growth in NA media: | The following 9 cultures were started to test bacteria growth in NA media: | ||
| - | * | + | *Negative Control (to make sure all strains grow in this minimal media) |
**E. coli K12 in Bushnell-Haas minimal media supplemented with glucose | **E. coli K12 in Bushnell-Haas minimal media supplemented with glucose | ||
| - | **P. putida | + | **P. putida oilsands in Bushnell-Haas minimal media supplemented with glucose |
| - | **P. flourescens | + | **P. flourescens oilsands in Bushnell-Haas minimal media supplemented with glucose |
*NA media | *NA media | ||
**E. coli K12 in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration) | **E. coli K12 in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration) | ||
| - | **P. putida | + | **P. putida oilsands in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration) |
| - | **P. flourescens | + | **P. flourescens oilsands in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration) |
*pH adjusted NA media | *pH adjusted NA media | ||
| - | **E. coli K12 in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration) pH adjusted between 8 and 9 | + | **E. coli K12 in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration), pH adjusted between 8 and 9 |
| - | **P. putida | + | **P. putida oilsands in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration), pH adjusted between 8 and 9 |
| - | **P. flourescens | + | **P. flourescens oilsands in Bushnell-Haas media supplemented with cyclohexane carboxylic acid (120 mg/L concentration), pH adjusted between 8 and 9 |
| - | 1 mL of the above media was added to a 15 mL falcon tube and inoculated with - | + | 1 mL of the above media was added to a 15 mL falcon tube and inoculated with -80°C freezer stock of each culture and grown in the 30C shaker (Psuedomonas strains are temperature sensitive). The cultures were started at 7:30 pm. |
==8/4/2010== | ==8/4/2010== | ||
| - | '''Culturing Bacteria in Naphthenic Acids ( | + | '''Culturing Bacteria in Naphthenic Acids (NAs)''' |
| - | At 12:30 pm the OD600 of the cultures started yesterday were measured and are listed below. These results must be taken with a grain of salt because the cultures were started from the - | + | At 12:30 pm the OD600 of the cultures started yesterday were measured and are listed below. These results must be taken with a grain of salt, because the cultures were started from the -80°C freezer stock with different amounts of inoculum: |
*P. putida BH-glu: 0.244 | *P. putida BH-glu: 0.244 | ||
| Line 182: | Line 184: | ||
*E. coli K12 BH-CHCA pH9: 0.0325 | *E. coli K12 BH-CHCA pH9: 0.0325 | ||
| - | P. flourescens | + | P. flourescens oilsands seems to love this medium and grew out very densely! P. putida (oilsands) showed signs of growing. E. coli K12 did not grow at all and pellets of dead cells appeared on the bottom of the culture flask. |
From these results we can start cultures for the biofilm assay experiment and time the cultures so they all become dense around the same day. If all cultures are started 24 hours in advance they all should saturate by the next day. | From these results we can start cultures for the biofilm assay experiment and time the cultures so they all become dense around the same day. If all cultures are started 24 hours in advance they all should saturate by the next day. | ||
| - | The E. coli K12 and P. putida oilsands cultures were placed back in the | + | The E. coli K12 and P. putida oilsands cultures were placed back in the 30°C shaker to keep growing. By 9:30pm, the P. putida oilsands cultures had saturated and the E. coli K12 cultures had still not grown. |
As a control for the biofilm assay E. coli K12 will be grown in M9 minimal media with glucose as a carbon source with and without casamino acids. We will not attempt to grow E. coli K12 in Bushnell-Haas media. | As a control for the biofilm assay E. coli K12 will be grown in M9 minimal media with glucose as a carbon source with and without casamino acids. We will not attempt to grow E. coli K12 in Bushnell-Haas media. | ||
| Line 193: | Line 195: | ||
'''Measuring pH of Bushnell-Haas minimal media with cyclohexane carboxylic acid adjusted to pH 9''' | '''Measuring pH of Bushnell-Haas minimal media with cyclohexane carboxylic acid adjusted to pH 9''' | ||
| - | Today Marc showed us how to use the pH meter in the Lin Lab. Yesterday the pH of the media was estimated by pH paper to be between 8 and 9. The actual pH of the media is 7.35. The pH of the already adjusted media from yesterday was increased by added more 250 uL of 0.1 M NaOH to 5mL of BH CHCA media and the pH | + | Today Marc showed us how to use the pH meter in the Lin Lab. Yesterday the pH of the media was estimated by pH paper to be between 8 and 9. The actual pH of the media is 7.35. The pH of the already adjusted media from yesterday was increased by added more 250 uL of 0.1 M NaOH to 5mL of BH-CHCA media and the pH read 7.6 using the pH probe. Another 250 uL of NaOH was added to this mixture and the pH read 8.8 using the pH probe. pH paper was dotted using this mixture to see the exact color it should turn when the pH is close to 9 (the paper looks more blue than green when the dot first appears). |
A more concentrated NaOH sterilized stock solution needs to be made so when NaOH is added to the media the salts are not diluted | A more concentrated NaOH sterilized stock solution needs to be made so when NaOH is added to the media the salts are not diluted | ||
| - | '''Bacterial Growth in pH | + | '''Bacterial Growth in pH 9''' |
| - | After determining that | + | After determining that 1 mL of the supposedly already adjusted BH-CHCA media needed 100 uL of 0.1 M NaOH solution per mL to really be close to a pH of 9, new overnight cultures were started of the two Pseudomonas oilsands strains in 1 mL of the BH-CHCA pH-adjusted media from yesterday with an additional 100 uL of 0.1 M NaOH added. These two cultures were placed in the 30°C shaker to grow out overnight at 8:30 pm. |
| - | '''Miniprep of pBAD take 2''' | + | '''Miniprep of pBAD, take 2''' |
| - | At 10:45 pm a 5 mL culture of the pBAD biobrick was started from the - | + | At 10:45 pm a 5 mL culture of the pBAD biobrick was started from the -80°C freezer stock in LB + 25 mg/mL KAN + 1 mM IPTG for a miniprep tomorrow morning. |
'''PCR of flu operon of E. coli K12''' | '''PCR of flu operon of E. coli K12''' | ||
| Line 213: | Line 215: | ||
PCR reagents were picked up from the life science store. | PCR reagents were picked up from the life science store. | ||
| - | * | + | *dNTPs |
| - | **These | + | **These dNTPs came in four separate containers individually in a 100 mM solution. The following recipe was used to make the 10 mM dNTP mixture: |
***100 uL of 100 mM dATP | ***100 uL of 100 mM dATP | ||
***100 uL of 100 mM dTTP | ***100 uL of 100 mM dTTP | ||
| Line 227: | Line 229: | ||
The PCR was run according to the following [[Media:8-4-2010_Colony_PCR_flu.pdf|colony PCR protocol]] | The PCR was run according to the following [[Media:8-4-2010_Colony_PCR_flu.pdf|colony PCR protocol]] | ||
| - | The final annealing cycle for 10 minutes at | + | The final annealing cycle for 10 minutes at 72°C was accidentally added in the last 30 cycle loop. This mistake was caught after 3 cycles and the PCR was stopped, the program fixed and the reaction restarted. |
'''Gel of flu colony PCR''' | '''Gel of flu colony PCR''' | ||
| - | The following gel was run in a 0.7% agarose gel at | + | The following gel was run in a 0.7% agarose gel at 85 V according to the protocol on the wiki: |
[[Image:8-4-2010_PCR_of_flu_operon.jpg|500px]] | [[Image:8-4-2010_PCR_of_flu_operon.jpg|500px]] | ||
| Line 241: | Line 243: | ||
'''Bacterial Growth at pH 9''' | '''Bacterial Growth at pH 9''' | ||
| - | At 9:00am the two cultures started in the BH-CHCA were tested to ensure the pH was around 9. Both P. putida oilsands and P. fluorescens oilsands showed slight signs of growth. Later, in the day at 3:30pm, the cultures | + | At 9:00am the two cultures started in the BH-CHCA were tested to ensure the pH was around 9. Both P. putida oilsands and P. fluorescens oilsands showed slight signs of growth. Later, in the day at 3:30pm, the cultures were denser; the OD600 of the culture was: |
*P. putida oilsands: 0.065 | *P. putida oilsands: 0.065 | ||
| Line 248: | Line 250: | ||
It should be noted some of the salts precipitated out of solution in the 1 mL cultures. | It should be noted some of the salts precipitated out of solution in the 1 mL cultures. | ||
| - | '''Miniprep of pBAD take 2''' | + | '''Miniprep of pBAD, take 2''' |
The miniprep was started at 8:45am. After 10 hours of growth the culture was saturated and the miniprep initialized according to the modified miniprep protocol. | The miniprep was started at 8:45am. After 10 hours of growth the culture was saturated and the miniprep initialized according to the modified miniprep protocol. | ||
| - | '''Nanodrop of pBAD take 2''' | + | '''Nanodrop of pBAD, take 2''' |
The concentration of the pBAD promoter miniprepped earlier today is 418 ng/uL. It is most likely still contaminated with this high concentration but we will still digest it and run a gel to check. | The concentration of the pBAD promoter miniprepped earlier today is 418 ng/uL. It is most likely still contaminated with this high concentration but we will still digest it and run a gel to check. | ||
| - | '''Digest of pBAD take 2''' | + | '''Digest of pBAD, take 2''' |
This digest was performed according to the digest protocol in the protocol section of the wiki: | This digest was performed according to the digest protocol in the protocol section of the wiki: | ||
| Line 265: | Line 267: | ||
'''PCR of flu operon''' | '''PCR of flu operon''' | ||
| - | The PCR of the flu operon was repeated 3 more times at higher annealing temperatures as this showed to have less unspecific product (shown by the gel from yesterday)according to this: [[Media:8-5-2010_Colony_PCR_flu.pdf|modified colony PCR protocol]]. The | + | The PCR of the flu operon was repeated 3 more times at higher annealing temperatures as this showed to have less unspecific product (shown by the gel from yesterday) according to this: [[Media:8-5-2010_Colony_PCR_flu.pdf|modified colony PCR protocol]]. The volumes of the reactions were increased to 50 uL for a PCR purification step afterwards. |
'''Gel of flu operon PCR and pBAD promoter digest''' | '''Gel of flu operon PCR and pBAD promoter digest''' | ||
| Line 407: | Line 409: | ||
A strong conclusion cannot be drawn about the biofilm forming abilities in minimal media with cyclohexane carboxylic acids (CHCA) as the cells did not grow very densely. This is the reason for the large error bars. | A strong conclusion cannot be drawn about the biofilm forming abilities in minimal media with cyclohexane carboxylic acids (CHCA) as the cells did not grow very densely. This is the reason for the large error bars. | ||
| - | It is interesting that P. fluorescens in pH 9 did not form biofilms even though the cell culture grew relatively well. We need a negative control to confirm this result. | + | It is interesting that P. fluorescens in pH 9 did not form biofilms, even though the cell culture grew relatively well. We need a negative control to confirm this result. |
==8/9/2010== | ==8/9/2010== | ||
| Line 415: | Line 417: | ||
Because there are unspecific products from the PCR of flu, a gel extraction will have to be used to isolate the desired DNA. | Because there are unspecific products from the PCR of flu, a gel extraction will have to be used to isolate the desired DNA. | ||
| - | 50 uL reaction volume for PCR was performed using 65.5C for the initial annealing temperature as shown in the following | + | 50 uL reaction volume for PCR was performed using 65.5C for the initial annealing temperature as shown in the following [[Media:8-9-2010_Colony_PCR_flu.pdf|colony PCR protocol for flu]]. The higher annealing temperature reduces unspecific products. |
'''Gel Extraction of flu operon''' | '''Gel Extraction of flu operon''' | ||
| Line 440: | Line 442: | ||
''Ann and Bryce'' | ''Ann and Bryce'' | ||
| - | '''Transformation of | + | '''Transformation of Constitutive Promoter and KAN backbone''' |
| - | The transformation was performed according to the electroporation protocol in the protocol section of the wiki | + | The transformation was performed according to the electroporation protocol in the protocol section of the wiki. |
The comp cell culture was started at 9:10am by adding the 8 mL overnight culture into 40mL of LB media in a 500 mL sterile flask. | The comp cell culture was started at 9:10am by adding the 8 mL overnight culture into 40mL of LB media in a 500 mL sterile flask. | ||
| Line 460: | Line 462: | ||
*backbone on LB+25 mg/mL KAN + 1mM IPTG | *backbone on LB+25 mg/mL KAN + 1mM IPTG | ||
| - | The plates were placed in the | + | The plates were placed in the 37°C incubator to grow out overnight. |
We plan on ligating the flu operon into the KAN backbone than cutting than inserting the promoter in front of this part. We must take this approach because the flu operon cannot be digested with PstI. | We plan on ligating the flu operon into the KAN backbone than cutting than inserting the promoter in front of this part. We must take this approach because the flu operon cannot be digested with PstI. | ||
| Line 493: | Line 495: | ||
*constitutive promoter: LB + 100 mg/mL AMP | *constitutive promoter: LB + 100 mg/mL AMP | ||
| - | These cultures were placed in the | + | These cultures were placed in the 37°C incubator to grow up. |
At 9:00pm, the cultures were miniprepped according to the modified miniprep protocol. | At 9:00pm, the cultures were miniprepped according to the modified miniprep protocol. | ||
| Line 515: | Line 517: | ||
'''Digest of constitutive promoter, KAN backbone and flu operon''' | '''Digest of constitutive promoter, KAN backbone and flu operon''' | ||
| - | The following digests were performed | + | The following digests were performed: |
*constitutive promoter: EcoRI and SpeI (labeled P ES) | *constitutive promoter: EcoRI and SpeI (labeled P ES) | ||
*constitutive promoter: EcoRI and XbaI (labeled P EX) | *constitutive promoter: EcoRI and XbaI (labeled P EX) | ||
| Line 526: | Line 528: | ||
Because the KAN backbone digested with EcoRI and SpeI is going to be PCR purified, double the amount of DNA was added and a 50 uL volume reaction was performed. | Because the KAN backbone digested with EcoRI and SpeI is going to be PCR purified, double the amount of DNA was added and a 50 uL volume reaction was performed. | ||
| - | Since no nanodrop reading was made for the flu operon, no water was added and only the gel extraction was used | + | Since no nanodrop reading was made for the flu operon, no water was added and only the gel extraction was used. |
'''Gel of constitutive promoter, KAN backbone and flu operon''' | '''Gel of constitutive promoter, KAN backbone and flu operon''' | ||
| Line 546: | Line 548: | ||
'''Ligation of flu operon with the KAN backbone''' | '''Ligation of flu operon with the KAN backbone''' | ||
| - | The ligation was performed according to the following [[Media:8-11-2010_Ligation_Calculations_for_flu_in_plasmid_pSB2K3.pdf|T4 ligase protocol]] | + | The ligation was performed according to the following [[Media:8-11-2010_Ligation_Calculations_for_flu_in_plasmid_pSB2K3.pdf|T4 ligase protocol]]. |
---- | ---- | ||
| Line 594: | Line 596: | ||
Attempted to perform electroporation according to [[Media:Electroporation Protocol 2.pdf|Electroporation Protocol 2]] with parts: | Attempted to perform electroporation according to [[Media:Electroporation Protocol 2.pdf|Electroporation Protocol 2]] with parts: | ||
| - | *pSB1AT3- Amp/Tet high copy backbone with RFP expression insert | + | *pSB1AT3 - Amp/Tet high copy backbone with RFP expression insert |
| - | *BBa_K145279- Tet resistance and Tet-activated GFP insert on Amp backbone | + | *BBa_K145279 - Tet resistance and Tet-activated GFP insert on Amp backbone |
| - | *BBa_I13504- Promoterless RBS-GFP insert on Amp backbone | + | *BBa_I13504 - Promoterless RBS-GFP insert on Amp backbone |
| - | *B0034- RBS on Amp backbone | + | *B0034 - RBS on Amp backbone |
However, cells took too long to grow out because only 1 mL of overnight was used to inoculate the 100 mL fresh culture. OD600 was only 0.142 after 4.5 hours. | However, cells took too long to grow out because only 1 mL of overnight was used to inoculate the 100 mL fresh culture. OD600 was only 0.142 after 4.5 hours. | ||
| Line 634: | Line 636: | ||
Parts: | Parts: | ||
| - | *pSB1AT3 (13A)- Amp/Tet high copy backbone with RFP expression insert | + | *pSB1AT3 (13A) - Amp/Tet high copy backbone with RFP expression insert |
| - | *BBa_K145279 (4L)- Tet resistance and Tet-activated GFP insert on Amp backbone | + | *BBa_K145279 (4L) - Tet resistance and Tet-activated GFP insert on Amp backbone |
| - | *BBa_I13504 (22L)- Promoterless RBS-GFP insert on Amp backbone | + | *BBa_I13504 (22L) - Promoterless RBS-GFP insert on Amp backbone |
| - | *B0034 (2M)- RBS on Amp backbone | + | *B0034 (2M) - RBS on Amp backbone |
| - | *O/N- 7:20 pm night before | + | *O/N - 7:20 pm night before |
| - | *Fresh culture- 11:45 am, grown out in Lin lab 30 degree shaker | + | *Fresh culture - 11:45 am, grown out in Lin lab 30 degree shaker |
*Cells harvested at 3 pm with OD of 0.419 | *Cells harvested at 3 pm with OD of 0.419 | ||
*1:100 dilution after wash step showed OD of 1.2 | *1:100 dilution after wash step showed OD of 1.2 | ||
| - | *Recovery was at | + | *Recovery was at 37°C for 1 hour |
*Undiluted, 1:10, and 1:100 concentrations were plated for each sample | *Undiluted, 1:10, and 1:100 concentrations were plated for each sample | ||
| Line 652: | Line 654: | ||
'''Gel of Flu + Kan Backbone Ligation''' | '''Gel of Flu + Kan Backbone Ligation''' | ||
| - | A Gel was | + | A Gel was run with the two digested ligations (from 8/30/2010) and failed to illuminate under UV light because of a mistake in the EtBr concentration used (2 mg/mL was used instead of the standard 10 mg/mL). |
| - | ''' Miniprep of Flu Ligation #1 & #2 (2nd Set) and 2M''' | + | '''Miniprep of Flu Ligation #1 & #2 (2nd Set) and 2M''' |
The Nanodrop values are as follows: | The Nanodrop values are as follows: | ||
| Line 792: | Line 794: | ||
*KT2440 inoculated into 2 mL LB in 15mL tube | *KT2440 inoculated into 2 mL LB in 15mL tube | ||
*others inoculated into 2 mL LB + 100μg/mL amp in 15mL tube | *others inoculated into 2 mL LB + 100μg/mL amp in 15mL tube | ||
| - | *all incubated in 30°C, 200rpm shaking | + | *all incubated in 30°C, 200rpm shaking - ERB 1230, 8:45pm |
Streaked KT2440 on LB plate from frozen | Streaked KT2440 on LB plate from frozen | ||
| - | *incubated 37°C | + | *incubated 37°C - ERB 1239 |
Added KT2440 to strain database | Added KT2440 to strain database | ||
Mixed M9 A and B solutions and diluted 1/50 in 200 mL sterile DIwater -- in 4 50mL tubes | Mixed M9 A and B solutions and diluted 1/50 in 200 mL sterile DIwater -- in 4 50mL tubes | ||
| Line 814: | Line 816: | ||
Repeated Flu PCR Gel from yesterday, with the following variations: | Repeated Flu PCR Gel from yesterday, with the following variations: | ||
*Regular amount of product/loading dye (4.8 and 1.2 uL) | *Regular amount of product/loading dye (4.8 and 1.2 uL) | ||
| - | * | + | *Run at 86 V for 60 min |
[[Image:29 Flu PCR Gel.jpg|400px]] | [[Image:29 Flu PCR Gel.jpg|400px]] | ||
| Line 824: | Line 826: | ||
Aliquot M9 and buffered M9 into five 15mL tubes each | Aliquot M9 and buffered M9 into five 15mL tubes each | ||
| + | |||
Need 15 mL stocks of each of the following media -- each 15 mL neutral and 15 mL buffered at pH 9: | Need 15 mL stocks of each of the following media -- each 15 mL neutral and 15 mL buffered at pH 9: | ||
*M9 | *M9 | ||
| Line 832: | Line 835: | ||
*LB | *LB | ||
Added 150 μL 40% glucose to appropriate tubes (.4% final concentration) | Added 150 μL 40% glucose to appropriate tubes (.4% final concentration) | ||
| + | |||
Aliquot 15 mL LB into each of two 15mL tubes | Aliquot 15 mL LB into each of two 15mL tubes | ||
| + | |||
Added NAs to appropriate tubes | Added NAs to appropriate tubes | ||
- Acros: (.25 mg/mL)*(15mL)/(.91 mg/μL) = '''4.12 μL''' | - Acros: (.25 mg/mL)*(15mL)/(.91 mg/μL) = '''4.12 μL''' | ||
| Line 867: | Line 872: | ||
Bryce came in in the evening and got the concentration of the purified samples, along with some minipreps according to the [[Media:Nanodrop_Protocol.pdf|Nanodrop]] protocol: | Bryce came in in the evening and got the concentration of the purified samples, along with some minipreps according to the [[Media:Nanodrop_Protocol.pdf|Nanodrop]] protocol: | ||
| - | Flu PCR Product- 29.8 ng/uL | + | Flu PCR Product - 29.8 ng/uL |
| - | 13A (pSB1AT3)- 240.5 ng/uL | + | 13A (pSB1AT3) - 240.5 ng/uL |
| - | 22L (BBa_I13504)- 467.4 ng/uL | + | 22L (BBa_I13504) - 467.4 ng/uL |
| - | 2M (B0034)- 661.4 ng/uL | + | 2M (B0034) - 661.4 ng/uL |
Bryce also digested the Flu operon and pSB1AT3 with EcoRI and SpeI according to the [[Media:General_DNA_Digest_Protocol.pdf|DNA Digest]] protocol. He used 17 uL of the Flu PCR product and 5 uL of pSB1AT3. | Bryce also digested the Flu operon and pSB1AT3 with EcoRI and SpeI according to the [[Media:General_DNA_Digest_Protocol.pdf|DNA Digest]] protocol. He used 17 uL of the Flu PCR product and 5 uL of pSB1AT3. | ||
Latest revision as of 01:36, 27 October 2010
| Sunday | Monday | Tuesday | Wednesday | Thursday | Friday | Saturday | |
| Week 6 | 8/1/2010 | 8/2/2010 | 8/3/2010 | 8/4/2010 | 8/5/2010 | 8/6/2010 | - |
| Week 7 | 8/8/2010 | 8/9/2010 | 8/10/2010 | 8/11/2010 | 8/12/2010 | - | 8/14/2010 |
| Week 8 | - | 8/16/2010 | - | - | - | - | - |
| Week 9 | - | - | - | - | 8/26/2010 | 8/27/2010 | - |
| Week 10 | 8/29/2010 | 8/30/2010 | 8/31/2010 | 9/1/2010 | 9/2/2010 | 9/3/2010 | 9/4/2010 |
| Week 11 | 9/5/2010 | - | - | - | - | - | - |
| Week 12 | 9/12/2010 | - | - | - | - | - | - |
| Week 13 | - | - | - | - | - | - | - |
| Week 14 | - | - | - | 9/29/2010 | 9/30/2010 | - | - |
Back to Oil Sands Main Notebook
8/1/2010
Nanodrop of pBAD and GFP verification
This miniprep was performed for the two cultures started last night according the the MODIFIED miniprep protocol. A frozen stock was made of the pBAD biobrick and placed in the -80°C freezer.
The DNA concentrations reported by the nanodrop are as follows:
- pBAD: 386 ng/uL (260/280=2.02 and 260/230=2.32)
- GFP: 25.8 ng/uL (260/280=2.22 and 260/230=2.72)
The pBAD grew out very densely; when IPTG is added, the copy number is over 100. The DNA concentration was very high.
Digest of pBAD and GFP verification
The parts were digested according to the protocol in the protocol section of the wiki for a 50 uL reaction volume:
- GFP #3 was digested with XbaI and PstI
- GFP #4 was digested with EcoRI and XbaI
- pBAD #1 was digested with EcoRI and SpeI
- pBAD #2 was digested with SpeI and PstI
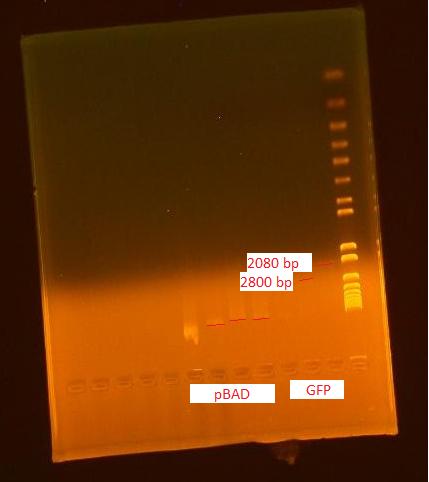
8/2/2010
Gel of Digested Parts
- Lane 1: Invitrogen 1 kb Plus ladder
- Lane 2: GFP #3 cut with XbaI and PstI (GFP: 720 bp; Backbone: 2079 bp)
- Lane 3: GFP #4 cut with EcoRI and XbaI
- Lane 4: uncut GFP plamid (only 1 uL left so did not show up on gel)
- Lane 5: pBAD cut with EcoRI and SpeI (pBAD: 1210 bp; Backbone: 4425 bp)
- Lane 6: pBAD cut with SpeI and PstI
- Lane 7: uncut pBAD (loaded very small amount into gel by accident)
- Lane 8: uncut pBAD (loaded 1 uL of miniprep into gel)
We did not get the pBAD part due to the miniprep culture overgrowing and the cells lysing their chromosomal DNA (resulting in extremely long bands in the gel). We will try miniprepping the DNA from this culture again letting the culture grow out for a shorter amount of time before the miniprep.
flu primers
Primers were ordered today to amplify the flu operon of E. coli K12 by colony PCR:
8/3/2010
Minimal Media
Minimal media was mixed today according to the following recipes (also found in the media recipe section of the notebooks):
Culturing Bacteria in Naphthenic Acids (NAs)
The following 9 cultures were started to test bacteria growth in NA media:
- Negative Control (to make sure all strains grow in this minimal media)
- E. coli K12 in Bushnell-Haas minimal media supplemented with glucose
- P. putida oilsands in Bushnell-Haas minimal media supplemented with glucose
- P. flourescens oilsands in Bushnell-Haas minimal media supplemented with glucose
- NA media
- E. coli K12 in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration)
- P. putida oilsands in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration)
- P. flourescens oilsands in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration)
- pH adjusted NA media
- E. coli K12 in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration), pH adjusted between 8 and 9
- P. putida oilsands in Bushnell-Haas minimal media supplemented with cyclohexane carboxylic acid (120 mg/L concentration), pH adjusted between 8 and 9
- P. flourescens oilsands in Bushnell-Haas media supplemented with cyclohexane carboxylic acid (120 mg/L concentration), pH adjusted between 8 and 9
1 mL of the above media was added to a 15 mL falcon tube and inoculated with -80°C freezer stock of each culture and grown in the 30C shaker (Psuedomonas strains are temperature sensitive). The cultures were started at 7:30 pm.
8/4/2010
Culturing Bacteria in Naphthenic Acids (NAs)
At 12:30 pm the OD600 of the cultures started yesterday were measured and are listed below. These results must be taken with a grain of salt, because the cultures were started from the -80°C freezer stock with different amounts of inoculum:
- P. putida BH-glu: 0.244
- P. putida BH-CHCA: 0.0245
- P. putida BH-CHCA pH 9: 0.033
- P. flourescens BH-glu: 0.396
- P. flourescens BH-CHCA: 0.219
- P. flourescens BH-CHCA pH9: 0.417
- E. coli K12 BH-glu: 0.017
- E. coli K12 BH-CHCA: 0.016
- E. coli K12 BH-CHCA pH9: 0.0325
P. flourescens oilsands seems to love this medium and grew out very densely! P. putida (oilsands) showed signs of growing. E. coli K12 did not grow at all and pellets of dead cells appeared on the bottom of the culture flask.
From these results we can start cultures for the biofilm assay experiment and time the cultures so they all become dense around the same day. If all cultures are started 24 hours in advance they all should saturate by the next day.
The E. coli K12 and P. putida oilsands cultures were placed back in the 30°C shaker to keep growing. By 9:30pm, the P. putida oilsands cultures had saturated and the E. coli K12 cultures had still not grown.
As a control for the biofilm assay E. coli K12 will be grown in M9 minimal media with glucose as a carbon source with and without casamino acids. We will not attempt to grow E. coli K12 in Bushnell-Haas media.
Measuring pH of Bushnell-Haas minimal media with cyclohexane carboxylic acid adjusted to pH 9
Today Marc showed us how to use the pH meter in the Lin Lab. Yesterday the pH of the media was estimated by pH paper to be between 8 and 9. The actual pH of the media is 7.35. The pH of the already adjusted media from yesterday was increased by added more 250 uL of 0.1 M NaOH to 5mL of BH-CHCA media and the pH read 7.6 using the pH probe. Another 250 uL of NaOH was added to this mixture and the pH read 8.8 using the pH probe. pH paper was dotted using this mixture to see the exact color it should turn when the pH is close to 9 (the paper looks more blue than green when the dot first appears).
A more concentrated NaOH sterilized stock solution needs to be made so when NaOH is added to the media the salts are not diluted
Bacterial Growth in pH 9
After determining that 1 mL of the supposedly already adjusted BH-CHCA media needed 100 uL of 0.1 M NaOH solution per mL to really be close to a pH of 9, new overnight cultures were started of the two Pseudomonas oilsands strains in 1 mL of the BH-CHCA pH-adjusted media from yesterday with an additional 100 uL of 0.1 M NaOH added. These two cultures were placed in the 30°C shaker to grow out overnight at 8:30 pm.
Miniprep of pBAD, take 2
At 10:45 pm a 5 mL culture of the pBAD biobrick was started from the -80°C freezer stock in LB + 25 mg/mL KAN + 1 mM IPTG for a miniprep tomorrow morning.
PCR of flu operon of E. coli K12
The primers ordered on Monday came in today!
PCR reagents were picked up from the life science store.
- dNTPs
- These dNTPs came in four separate containers individually in a 100 mM solution. The following recipe was used to make the 10 mM dNTP mixture:
- 100 uL of 100 mM dATP
- 100 uL of 100 mM dTTP
- 100 uL of 100 mM dGTP
- 100 uL of 100 mM dCTP
- 600 uL of ultra pure water
- These dNTPs came in four separate containers individually in a 100 mM solution. The following recipe was used to make the 10 mM dNTP mixture:
- Phusion High Fidelity Polymerase:
- comes with buffer
- used to amplify long pieces of DNA
- 50 rxn PCR purification kit
The PCR was run according to the following colony PCR protocol
The final annealing cycle for 10 minutes at 72°C was accidentally added in the last 30 cycle loop. This mistake was caught after 3 cycles and the PCR was stopped, the program fixed and the reaction restarted.
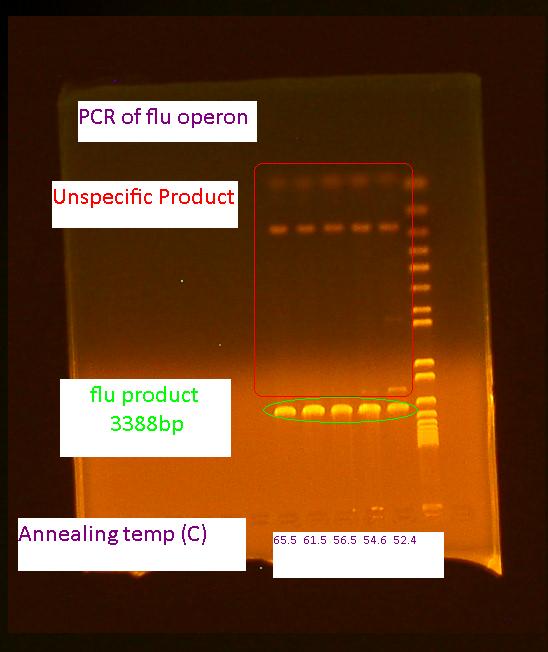
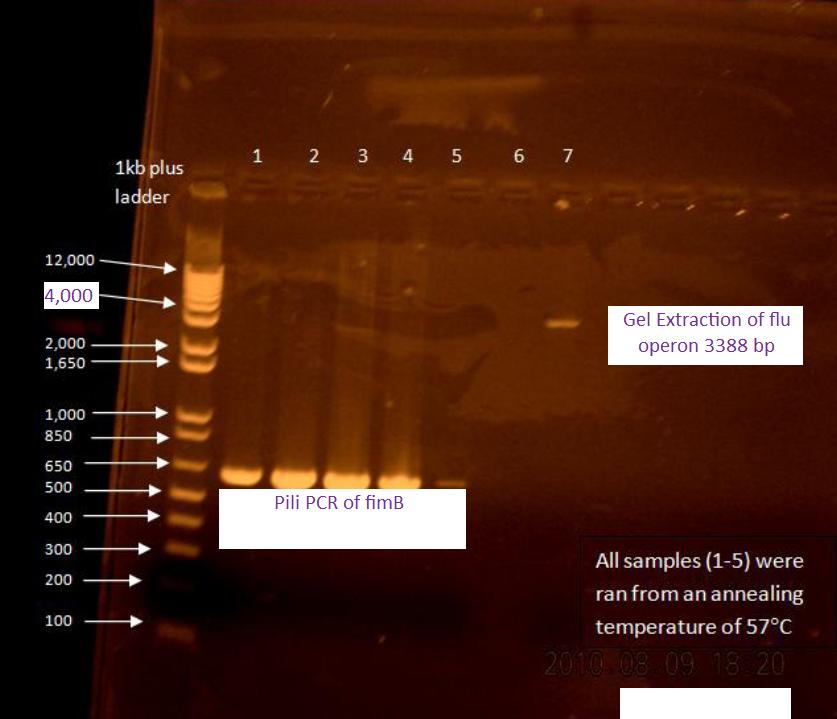
Gel of flu colony PCR
The following gel was run in a 0.7% agarose gel at 85 V according to the protocol on the wiki:
For the first 5 cycles the annealing temperature was lowered to the Tm of the 16 or 18 bp that was complementary to the E. coli K12 template. A gradient of temperature was run for the annealing temperatures of the first five cycles as labeled in the gel.
8/5/2010
Bacterial Growth at pH 9
At 9:00am the two cultures started in the BH-CHCA were tested to ensure the pH was around 9. Both P. putida oilsands and P. fluorescens oilsands showed slight signs of growth. Later, in the day at 3:30pm, the cultures were denser; the OD600 of the culture was:
- P. putida oilsands: 0.065
- P. fluorescens oilsands: 0.433
It should be noted some of the salts precipitated out of solution in the 1 mL cultures.
Miniprep of pBAD, take 2
The miniprep was started at 8:45am. After 10 hours of growth the culture was saturated and the miniprep initialized according to the modified miniprep protocol.
Nanodrop of pBAD, take 2
The concentration of the pBAD promoter miniprepped earlier today is 418 ng/uL. It is most likely still contaminated with this high concentration but we will still digest it and run a gel to check.
Digest of pBAD, take 2
This digest was performed according to the digest protocol in the protocol section of the wiki:
pBAD #3 was digested with EcoRI and SpeI pBAD #4 was digested with SpeI and PstI
PCR of flu operon
The PCR of the flu operon was repeated 3 more times at higher annealing temperatures as this showed to have less unspecific product (shown by the gel from yesterday) according to this: modified colony PCR protocol. The volumes of the reactions were increased to 50 uL for a PCR purification step afterwards.
Gel of flu operon PCR and pBAD promoter digest
The gel ran exactly the same for the pBAD promoter as on 8/2/2010. The sample is probably contaminated and a new colony should be picked from the transformation plate and screened.
Also, no bands appeared for the flu PCR. One error might be because we did not include an initial cell lysis step. The PCR will be attempted again tomorrow.
Biofilm assay in minimal media
Cultures were started according to the biofilm assay protocol for the inoculation of the microplate; these will be used in the biofilm assay tomorrow.
8/6/2010
PCR of flu take 3
The first PCR of the flu operon was repeated with an added cell lysis step according to the following modified protocol:
Gel of PCR of flu take 3
Invitrogen 1 kb plus ladder
Due to the unspecific product the flu operon will be purified with gel purification.
Biofilm Assay
The OD600 of the cultures started yesterday are listed below along with the volume for resuspension after 1 mL of culture was pelleted:
| OD600 | Volume for resuspension (uL) | |
| E. coli K12 LB | 0.914 | 228 |
| P. putida KT2440 LB | 1.168 | 292 |
| P. putida oilsands LB | 1.119 | 280 |
| P. fluorescens oilsands LB | 1.283 | 321 |
| E. coli K12 M9 minimal media plus glucose | 0.391 | 98 |
| P. putida KT2440 Bushnell-Hass media plus glucose | 0.545 | 136 |
| P. putida oilsands Bushnell-Hass media plus glucose | 0.385 | 96 |
| P. fluorescens oilsands Bushnell-Hass media plus glucose | 0.504 | 126 |
| E. coli K12 M9 minimal media plus glucose plus casamino acids | 0.814 | 204 |
| P. putida KT2440 Bushnell-Hass media plus glucose plus casamino acids | 1.185 | 296 |
| P. putida oilsands Bushnell-Hass media plus glucose plus casamino acids | 1.121 | 280 |
| P. fluorescens oilsands Bushnell-Hass media plus glucose plus casamino acids | 1.191 | 298 |
| P. putida KT2440 Bushnell-Hass media plus cyclohexane carboxylic acid | 0.0345 | 8.6 |
| P. putida oilsands Bushnell-Hass media plus cyclohexane carboxylic acid | 0.144 | 36 |
| P. fluorescens oilsands Bushnell-Hass media plus cyclohexane carboxylic acid | 0.197 | 49 |
| P. putida KT2440 Bushnell-Hass media plus cyclohexane carboxylic acid pH9 | 0.034 | 8.5 |
| P. putida oilsands Bushnell-Hass media plus cyclohexane carboxylic acid pH9 | 0.002 | 0.06 |
| P. fluorescens oilsands Bushnell-Hass media plus cyclohexane carboxylic acid pH9 | 0.199 | 49.8 |
Not all of the cultures grew out (P. putida KT2440 Bushnell-Haas media plus cyclohexane carboylic acid, P. putida KT2440 Bushnell-Haas media plus cyclohexane carboylic acid pH 9, P. putida oilsands Bushnell-Haas media plus cyclohexane carboxylic acid pH 9). The modified bio assay protocol was used to inoculate the microplate.
The microplate was inoculated at 4:30pm.
8/8/2010
Biofilm Assay
The washing and CV staining of the bioassay was performed according to the modified protocol on 8/6/2010.
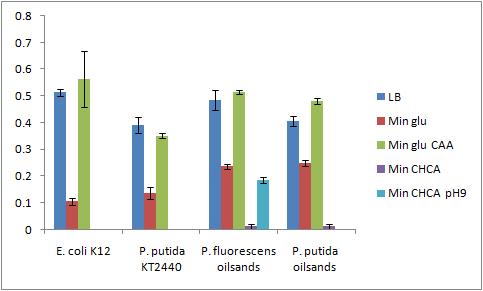
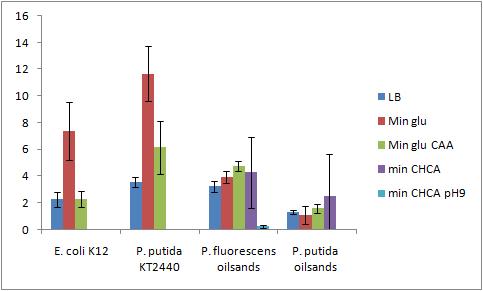
The OD 600 of the plate after the bioassay growth is shown in the following graph:
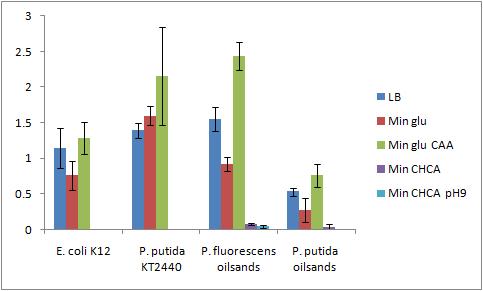
The Crystal Violet OD 600 after staining is show in the following graph:
The Crystal Violet OD 600 to OD 600 after growth ratio is shown in the following graph:
The negative control (E. coli K12 in M9 minimal media with glucose) grew one of the best biofilms; this contradicted what was expected. From here we need to find a knock out strain that is deficient in forming biofilms.
A strong conclusion cannot be drawn about the biofilm forming abilities in minimal media with cyclohexane carboxylic acids (CHCA) as the cells did not grow very densely. This is the reason for the large error bars.
It is interesting that P. fluorescens in pH 9 did not form biofilms, even though the cell culture grew relatively well. We need a negative control to confirm this result.
8/9/2010
PCR of flu for gel extraction
Because there are unspecific products from the PCR of flu, a gel extraction will have to be used to isolate the desired DNA.
50 uL reaction volume for PCR was performed using 65.5C for the initial annealing temperature as shown in the following colony PCR protocol for flu. The higher annealing temperature reduces unspecific products.
Gel Extraction of flu operon
A gel extraction was performed according to the following gel extraction protocol.
20 uL of the flu #14 PCR was mixed with 5 uL of blue juice to run the gel.
The weight of the eppendorf tube was 1.0284 g.
The weight of the eppendorf tube with the sample was 1.2030 g.
The weight of the gel sample was 0.175 g.
524 uL of QG buffer was added to dissolve the gel sample.
No DNA appeared on the nanodrop. Marc ran the flu gel purified sample on his gel to confirm no DNA eluted.
DNA did appear on the gel that Marc ran so we do have DNA from the gel extraction. The nanodrop may not have been able to pick it up because it was too low of a concentration or it was contaminated (very high 260/230 reading).
8/10/2010
Ann and Bryce
Transformation of Constitutive Promoter and KAN backbone
The transformation was performed according to the electroporation protocol in the protocol section of the wiki.
The comp cell culture was started at 9:10am by adding the 8 mL overnight culture into 40mL of LB media in a 500 mL sterile flask.
The OD600 of the culture was checked at 11:20am and was 1.0. This culture was slightly overgrown but was immediately placed on ice and used anyways (this might lower the efficiency of the procedure).
The biobricks we are transforming today are:
- J23100 (constitutive promoter)
- I14018 (PCR purify to remove promoter and be left with backbone that has KAN resistance)
- (pBAD biobrick taken from the registry as a positive control)
The transformation was done at 2:15pm and the cells were plated on the following plates:
- negative control (comp cells only) on LB+100 mg/mL AMP
- constitutive promoter on LB+100 mg/mL AMP
- negative control (comp cells only) on LB+25 mg/mL KAN + 1mM IPTG
- positive control (pBAD promoter taken from the registry) on LB+25 mg/mL KAN + 1mM IPTG
- backbone on LB+25 mg/mL KAN + 1mM IPTG
The plates were placed in the 37°C incubator to grow out overnight.
We plan on ligating the flu operon into the KAN backbone than cutting than inserting the promoter in front of this part. We must take this approach because the flu operon cannot be digested with PstI.
Marcus
Brought over 5 mL of each standard pH buffer (4, 7, 10) from Lin lab.
Started NA extraction from oil sand ore:
- Made 1M NaOH solution in Lin lab
- 1.969 g NaOH pellets
- 50 mL dH2O
- Mixed ore with 1M NaOH in 600 mL beaker
- 51.3 g 1M NaOH
- 38.6 g ore
- Put beaker in shaker for 48 hours
See the NA Extraction Protocol.
8/11/2010
Miniprep of constitutive promoter and backbone with KAN resistance
The transformation was successful; both plates with the constitutive promoter and the backbone with KAN resistance grew up (even though the positive control did not grow...).
The constitutive promoter part is followed by RFP. We saw this on the transformation plates because the colonies were red.
A colony was picked from each plate and at 9:00am and 5mL cultures were started in the following media:
- backbone: LB + 50 mg/mL KAN + 1mM IPTG
- constitutive promoter: LB + 100 mg/mL AMP
These cultures were placed in the 37°C incubator to grow up.
At 9:00pm, the cultures were miniprepped according to the modified miniprep protocol.
Nanodrop of constitutive promoter and KAN backbone
The concentration of the minipreps are as follows:
- backbone: 60 ng/uL (260/280= 1.87, 260/230= 2.06)
- constitutive promoter: 200ng/uL (260/280= 1.89, 260/230= 2.30)
PCR of flu operon from the gel extraction product
Using gel extraction product for ligations lowers the efficiency of the ligation. To get around this problem we performed another PCR reaction using the gel extraction product as the template according to the following PCR protocol.
Gel of PCR of flu operon from gel extraction product
There was more unspecific product from trying the PCR this reaction from the gel extracted part, so the gel extracted part will be used for further genetic manipulations.
8/12/2010
Digest of constitutive promoter, KAN backbone and flu operon
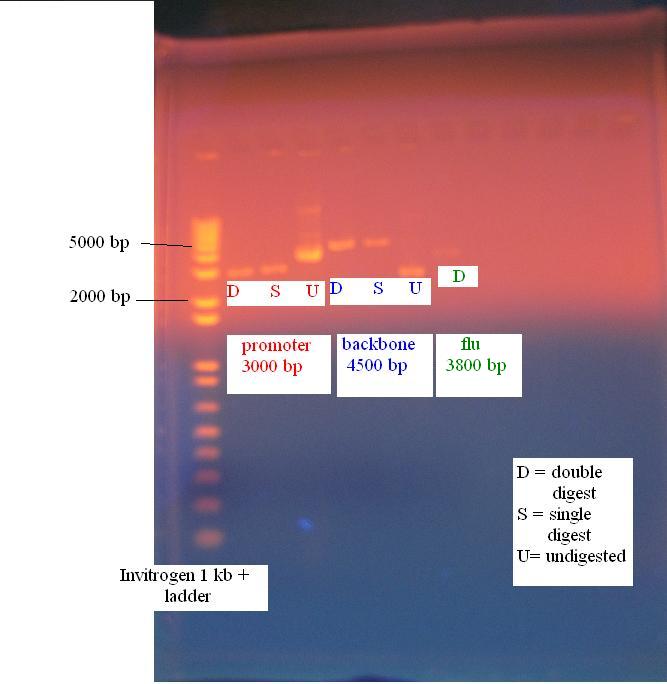
The following digests were performed:
- constitutive promoter: EcoRI and SpeI (labeled P ES)
- constitutive promoter: EcoRI and XbaI (labeled P EX)
- KAN backbone: EcoRI and SpeI (labeled B ES)
- KAN backbone: SpeI and PstI (labeled B SP)
- flu operon (from gel purified product): EcoRI and SpeI (labeled F ES)
All digests except for the KAN backbone digested with EcoRI and SpeI were 20 uL volumes. 1 uL of each enzyme was added along with 0.5 uL of BSA to the 20 uL reactions and the rest of the reagents were ajusted by multiplying the 50 uL volume by 0.4. To make up for the extra volume of BSA and enzymes, the extra amount was subtracted from the amount of water added.
Because the KAN backbone digested with EcoRI and SpeI is going to be PCR purified, double the amount of DNA was added and a 50 uL volume reaction was performed.
Since no nanodrop reading was made for the flu operon, no water was added and only the gel extraction was used.
Gel of constitutive promoter, KAN backbone and flu operon
All parts digested accordingly and are ready for the ligation.
PCR purification of KAN backbone
The PCR purification was performed with the 50 uL digest of the KAN backbone with EcoRI and SpeI according to the protocol in the protocol section of the wiki.
250 uL of buffer PB was used to mix with the digest before adding the mixture to the column.
Nanodrop of PCR purified KAN backbone
The PCR purified KAN backbone had a concentration of 14.8 ng/uL with a 260/280 ratio of 1.86 and a 260/230 ratio of 0.91. These ratios indicate that the sample is contaminated but there is still a high enough peak at 260 to show DNA is present. Another gel could be run to verify this, but this step will be skipped and the DNA will be used for the ligation.
Ligation of flu operon with the KAN backbone
The ligation was performed according to the following T4 ligase protocol.
Marcus
Removed ore/NaOH extraction mixture from shaker. Poured liquid into 50 mL tube, and strained the solids through a coffee filter and funnel to collect as much liquid as possible; 20mL was recovered. This was filtered through a 50 mL Steriflip vacuum tube; however, the filter became clogged and a second one was used. This required an extended vacuum time (in the future the mixture should be centrifuged first for several minutes). Recovered a total of about 40 mL (???).
8/13/2010
Marcus
- Autoclaved 1.5 mL tubes.
- Took two bags of autoclave waste to DOW building to be autoclaved.
8/14/2010
Transformation of the Flu Operon + Kan backbone ligation
The Heat shock transformation protocol was used.
E.Coli DH5α was used as competent cells and had a prewashing OD600 of .845 and a post washing OD600 of .787.
Two separate transformations were performed both with 100 uL Comp. Cells + 10 uL Flu + KAN BB Ligation.
Both transformations were plated on LB+Kan+IPTG(30 uL) and are currently incubating at 37 C in the ERB.
8/16/2010
Transformation of Flu Operon + Kanamycin Backbone Ligation
- The first transformation plate showed no colonies.
- The second transformation plate showed minimal colonies.
Both plates were left to incubate longer as the slow growth may be due to the high concentration of Kanamycin (50mg/mL) added to the plates.
8/26/2010
Overnight Cultures of the Flu Operon + Kanamycin Backbone Transformation
Colonies appeared on both transformation plates.
Overnight cultures were started from both plates in 5 mL LB + KAN 50 + 1 mM IPTG.
Marcus
Attempted to perform electroporation according to Electroporation Protocol 2 with parts:
- pSB1AT3 - Amp/Tet high copy backbone with RFP expression insert
- BBa_K145279 - Tet resistance and Tet-activated GFP insert on Amp backbone
- BBa_I13504 - Promoterless RBS-GFP insert on Amp backbone
- B0034 - RBS on Amp backbone
However, cells took too long to grow out because only 1 mL of overnight was used to inoculate the 100 mL fresh culture. OD600 was only 0.142 after 4.5 hours.
Made 10 Amp and 10 LB plates.
8/27/2010
Overnight Cultures of the Flu Operon + Kanamycin Backbone Transformation Take 2
The first set of overnight cultures are experiencing slow growth.
A second set of overnight cultures were started with lower Kanamycin levels (5 mL LB + KAN 25 + 1 mM IPTG).
8/29/2010
Miniprep & Nanodrop of Flu + Kan Backbone Ligation
Before the miniprep .5 mL of both cultures were used to make a frozen stock. The frozen stocks now reside in the -20 C freezer in the ERB.
The first set of overnight cultures were miniprepped and eluted in EB Buffer.
After Nanodropping the DNA Concentrations for plated #1 and #2 were 9.5 and 4.0 ng/uL, respectively.
8/30/2010
Digest of Flu + Kan Backbone Ligation
The miniprepped cells (from 8/29/2010) were digested with EcoRI & Spel to screen if the ligation worked properly or if mixed sites formed. 20 uL of DNA from ligation #1 and 25 uL of DNA from ligation #2 were used for the digest to compensate for the low miniprep yields.
Marcus and John
Repeated electroporation attempted on 8/26 using Electroporation Protocol 2.
Parts:
- pSB1AT3 (13A) - Amp/Tet high copy backbone with RFP expression insert
- BBa_K145279 (4L) - Tet resistance and Tet-activated GFP insert on Amp backbone
- BBa_I13504 (22L) - Promoterless RBS-GFP insert on Amp backbone
- B0034 (2M) - RBS on Amp backbone
- O/N - 7:20 pm night before
- Fresh culture - 11:45 am, grown out in Lin lab 30 degree shaker
- Cells harvested at 3 pm with OD of 0.419
- 1:100 dilution after wash step showed OD of 1.2
- Recovery was at 37°C for 1 hour
- Undiluted, 1:10, and 1:100 concentrations were plated for each sample
We didn't quite have enough comp cells because we forgot to account for controls when making the cells. A couple samples were a bit short (22L and another).
8/31/2010
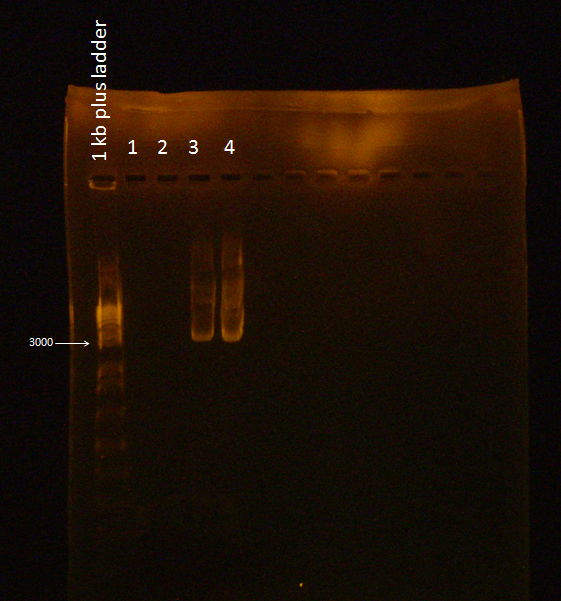
Gel of Flu + Kan Backbone Ligation
A Gel was run with the two digested ligations (from 8/30/2010) and failed to illuminate under UV light because of a mistake in the EtBr concentration used (2 mg/mL was used instead of the standard 10 mg/mL).
Miniprep of Flu Ligation #1 & #2 (2nd Set) and 2M
The Nanodrop values are as follows:
- Flu #1 - 23.2 ng/uL
- Flu #2 - 18.8 ng/uL
- 2M - 17.8 ng/uL
9/1/2010
Gel of Flu + Kan Backbone Ligation (Take 2)
A Gel was ran with the same two digested ligations (from 8/30/2010) and failed to illuminate under UV light. The source of error is believed to be that the DNA ran off the gel because the voltage was originally set at 85V (to run for 1.5 hrs) and upon returning (after 1.5 hrs) the voltage had been changed to 105V (by another lab member roughly 15 minutes into the start of the gel).
9/2/2010
Digest of Flu + Kan Backbone Ligation (Take 2)
The miniprepped cells (from 8/31/2010) were digested with Xba1 (To screen if the ligation worked by checking for a band at 7790 base pairs). Ann's digest calculator was used to calculate the proper amount of DNA to use.
Miniprep of 22L, 13A, and Kan Backbone
The DNA concentrations from the Miniprep by Nanodropping are as follows:
- 22L - 183.7 ng/uL
- 13A - 54.7 ng/uL
- KAN BB - 188.1 ng/uL
9/3/2010
Overnight cultures of Flu Ligation 1 & 2 (set 3), KAN BB, 2M
The Flu ligations and KAN Backbone cultures were started in 5 mL of LB + KAN 25 + 1 mM IPTG.
The 2M culture was started in 5 mL of LB + AMP.
9/4/2010
Miniprep of of Flu Ligation 1 & 2 (set 3), KAN BB, 2M
The DNA concentrations from the Miniprep by Nanodropping are as follows:
- Flu #1 - 24.1 ng/uL
- Flu #2 - 22.8 ng/uL
- 2M - 44.0 ng/uL
- KAN BB - 24.4 ng/uL
9/5/2010
Gel of Flu Ligation Transformation (take 3)
Marcus
Ore extracts were combined into a single tube and re-filtered, then stored in acid storage cabinet.
9/11/2010
Marcus and John
Made dilution protocol for CHCA starting with 1500 mg/L stock solution made by Ann previously. Made 500, 250, 125, 50, 10, 5, and 1 mg/L solutions, with at least 0.75 mL left of each after dilutions. These were stored in the fume hood and then given to Mike for analysis on the HPLC.
Also, liquid waste was autoclaved.
9/12/2010
Alex
Made E. coli K12 broth culture from plate
- 2 mL LB in 15mL tube
- 37°C, 200 rpm shaking - ERB 1230
Uploaded NanR cloning protocol
9/24/2010
Marcus, Bryce, and Ben
Ran PCR of Flu operon according to the NanR cloning PCR protocol, with modifications as follows:
- 1x PCR Mix:
- 36.25 uL Ultra pure H2O
- 10 uL 5x Phusion Buffer
- 1 uL 10 mM dNTPs
- 0.625 uL Forward Flu Primer
- 0.625 uL Reverse Flu Primer
- 1 uL DNA Template (1:10 O/N dilution)
- 0.5 uL Phusion polymerase
- PCR Program (FLUPCR)
- 1. 96 C for 6 min
- 2. 98 C for 10 sec
- 3. 65.5 C for 30 sec
- 4. 72 C for 1 min 45 sec
- 5. Return to step 2 four times
- 6. 98 C for 10 sec
- 7. 67 C for 30 sec
- 8. 72 C for 1 min 45 sec
- 9. Return to step six 29 times
- 10. 72 C for 10 min
- 11. 4 C store
Unfortunately, a math mistake was made while making the master mix, which was discovered due to having too much master mix left over. The volumes used to create the master mix were:
- 181.25 uL Ultra pure H2O
- 100 uL 5x Phusion Buffer
- 10 uL 10 mM dNTPs
- 6.25 uL Forward Flu Primer
- 6.25 uL Reverse Flu Primer
- 10 uL DNA Template
2.5 uL Phusion polymerase
9/27/2010
Marcus
Got some chloramphenicol from the Lin lab (34 mg/mL 1000x stock) and made 10 chloramphenicol plates and 10 LB plates. Also, made 45 mL of a LB+CM34 stock, and autoclaved 500 mL of LB.
9/28/2010
Marcus
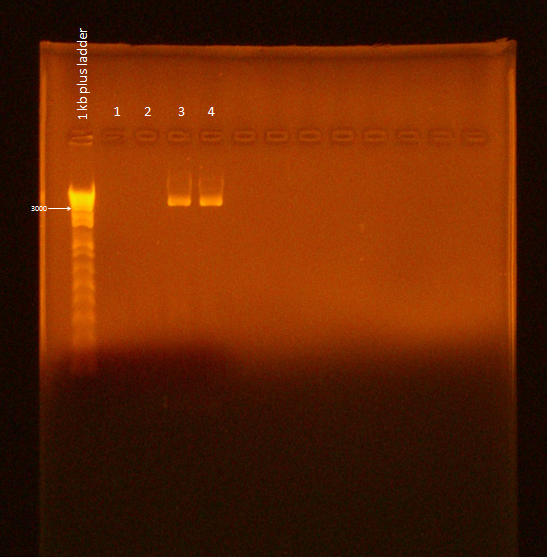
Ran gel of Flu PCR product. Used the Gel Electrophoresis protocol with the following variations:
- Doubled PCR product volume, in case gel extraction was needed
- 9.6 uL PCR product, 2.4 uL Blue Juice
- Ran at 110V for 50 min
It appears samples 3 and 4 were successful. However, the gel needs to be re-run because of excessive smearing. Jeremy Minty suggested lower voltage and less time to remedy this. Also, "nonspecific" bands seen by Ann and Bryce were nearly nonexistent and also seen in lanes 1 and 2, suggesting that they are not nonspecific product, but something else, such as primer-dimer.
9/29/2010
Alex and John
Dissolved M9 salts to 50X
- All of component A in 1000 mL DIwater
- All of component B in 1000 mL DIwater
Autoclaved both: each split into two 1000mL bottles filled w/ 500mL each (four bottles total)
- 30 min sterilize time
Began biofilm assay following Alex's biofilm assay protocol
Made broth cultures of P. putida LD1, P. fluorescens LD2, and LD1+LD2 coculture from plates, and P. putida KT2440 from Lin Lab frozen stock
- KT2440 inoculated into 2 mL LB in 15mL tube
- others inoculated into 2 mL LB + 100μg/mL amp in 15mL tube
- all incubated in 30°C, 200rpm shaking - ERB 1230, 8:45pm
Streaked KT2440 on LB plate from frozen
- incubated 37°C - ERB 1239
Added KT2440 to strain database Mixed M9 A and B solutions and diluted 1/50 in 200 mL sterile DIwater -- in 4 50mL tubes
- each tube 1 mL M9A, 1 mL M9B and 48 mL water
Alex
Uploaded Alex's biofilm assay protocol
Added borax to two of the M9 tubes (100 mL) to buffer at ~pH 9.2
- (.01 mol/L)*(.05 L)*(381.37 g/mol) = 190.7 mg borax/50 mL water
- Tested pH with pH indicator strips
- ~9
- Vacuum-filtered 100 mL w/ steriflips
Marcus
Repeated Flu PCR Gel from yesterday, with the following variations:
- Regular amount of product/loading dye (4.8 and 1.2 uL)
- Run at 86 V for 60 min
Results look much better, bands are tighter, and conclusions are the same as yesterday. No gel extraction will be necessary.
9/30/2010
Alex
Aliquot M9 and buffered M9 into five 15mL tubes each
Need 15 mL stocks of each of the following media -- each 15 mL neutral and 15 mL buffered at pH 9:
- M9
- M9 + 0.4% glucose
- M9 + Acros NAs (250mg/L)
- M9 + Sigma Aldrich NAs (250mg/L)
- M9 + both Acros & Sigma NAs (each 250mg/L)
- LB
Added 150 μL 40% glucose to appropriate tubes (.4% final concentration)
Aliquot 15 mL LB into each of two 15mL tubes
Added NAs to appropriate tubes
- Acros: (.25 mg/mL)*(15mL)/(.91 mg/μL) = 4.12 μL - Sigma: (.25 mg/mL)*(15mL)/(.92 mg/μL) = 4.08 μL
- added 4.1 μL of each NA to each respective tube
Borax was added to one tube of LB to buffer at pH9:
- (190.7 mg)*(15μL/50μL) = 57.2 mg per 15mL LB
- tested pH with indicator strip
- ~9
Tested pH of neutral M9 with indicator strip
- ~7
Moved KT2440 LB plate from 37°C to 4°C -- ERB 1239
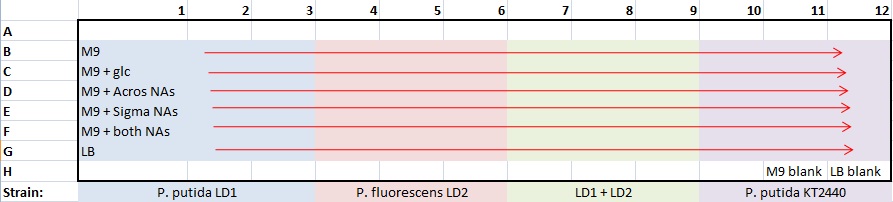
Continued Alex's biofilm assay protocol
- made one 96-well plate for all strains and media at pH7 only
- plate template :
- 100 μL/well: 99 μL medium + 1 μL culture
- covered plate with gas-permeable membrane
- Started 24-hr kinetic reading in Lin Lab microplate reader
- 30°C
- OD600 absorbance
- read every 30 min
Made broth cultures for LD1, LD2 and KT2440 from yesterday's overnight broths
- 2 mL each in a 15mL tube
- 30°C, 200 rpm shaking - 6:20pm
Discarded yesterday's overnight broths
Marcus
Purified Flu PCR product with Qiagen PCR purification kit and eluted in ultra pure H2O instead of EB. 35 uL remaining from samples 3 and 4 of PCR were combined and purified, and stored at 4 C.
Bryce came in in the evening and got the concentration of the purified samples, along with some minipreps according to the Nanodrop protocol:
Flu PCR Product - 29.8 ng/uL 13A (pSB1AT3) - 240.5 ng/uL 22L (BBa_I13504) - 467.4 ng/uL 2M (B0034) - 661.4 ng/uL
Bryce also digested the Flu operon and pSB1AT3 with EcoRI and SpeI according to the DNA Digest protocol. He used 17 uL of the Flu PCR product and 5 uL of pSB1AT3.
 "
"