Team:Johns Hopkins/Project
From 2010.igem.org
(→Instrumentation) |
(→Quantitative Characterization of FKS2 CDRE and PMC1 CDRE in Vacuole Positive and Negative Yeast) |
||
| (83 intermediate revisions not shown) | |||
| Line 17: | Line 17: | ||
{ | { | ||
display:block; | display:block; | ||
| - | width: | + | width:112px; |
font-weight:bold; | font-weight:bold; | ||
color:#FFFFFF; | color:#FFFFFF; | ||
| Line 39: | Line 39: | ||
<li><a href="https://2010.igem.org/Team:Johns_Hopkins/Modeling">Modeling</a></li> | <li><a href="https://2010.igem.org/Team:Johns_Hopkins/Modeling">Modeling</a></li> | ||
<li><a href="https://2010.igem.org/Team:Johns_Hopkins/Parts">Parts</a></li> | <li><a href="https://2010.igem.org/Team:Johns_Hopkins/Parts">Parts</a></li> | ||
| + | <li><a href="https://2010.igem.org/Team:Johns_Hopkins/Device">Device</a></li> | ||
<li><a href="https://2010.igem.org/Team:Johns_Hopkins/Notebook">Notebook</a></li> | <li><a href="https://2010.igem.org/Team:Johns_Hopkins/Notebook">Notebook</a></li> | ||
<li><a href="https://2010.igem.org/Team:Johns_Hopkins/Safety">Safety</a></li> | <li><a href="https://2010.igem.org/Team:Johns_Hopkins/Safety">Safety</a></li> | ||
| Line 45: | Line 46: | ||
</html> | </html> | ||
== '''Abstract''' == | == '''Abstract''' == | ||
| - | If the goal of iGEM and the Parts Registry is to take the messy world of genetic engineering and transform it into something like the standardized world of electrical engineering, it may be useful if electronic systems could directly interface with biological systems. Past iGEM projects have used chemical or optical stimuli to actuate transcriptional responses. Our project, however, seeks to add voltage sensitivity to Saccharomyces cerevisiae. Baker’s yeast was chosen because in some sense yeast have a system that responds to voltage input. With a voltage stimulus one can open the voltage-gated calcium channels of yeast, causing calcium ions to rush into the cytoplasm. This causes calcineurin to dephosphorylate Crz1, which enters the nucleus and binds various promoters. Our group presents a library of characterized Crz1-sensitive | + | If the goal of iGEM and the Parts Registry is to take the messy world of genetic engineering and transform it into something like the standardized world of electrical engineering, it may be useful if electronic systems could directly interface with biological systems. Past iGEM projects have used chemical or optical stimuli to actuate transcriptional responses. Our project, however, seeks to add voltage sensitivity to ''Saccharomyces cerevisiae''. Baker’s yeast was chosen because in some sense yeast have a system that responds to voltage input. With a voltage stimulus one can open the voltage-gated calcium channels of yeast, causing calcium ions to rush into the cytoplasm. This causes calcineurin to dephosphorylate Crz1, which enters the nucleus and binds various promoters. Our group presents a library of characterized Crz1-sensitive binding sites of both naturally-occurring and synthetic varieties that can be integrated into promoters. Genes downstream of these promoters are thus voltage-regulated in media containing calcium. |
| + | |||
| + | <br> | ||
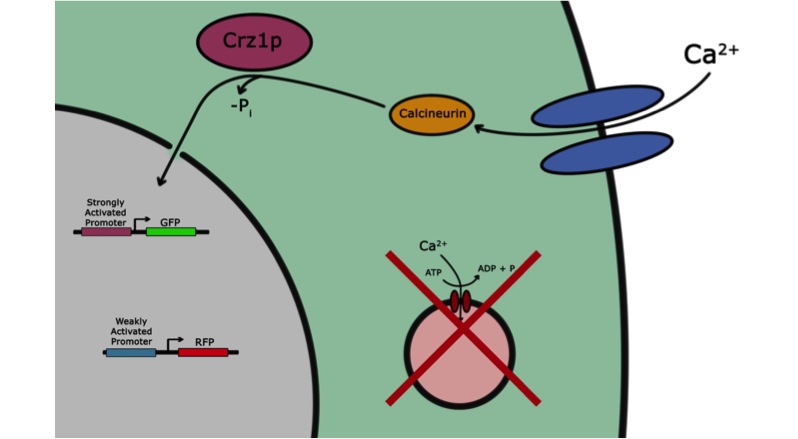
| + | [[Image:plan.jpg|1000px|center|frame|'''The Plan, Visualized:''' In this image, the pathway we are utilizing is schematically represented in an extremely accurate fashion by blobs and arrows. The calcium channels (mid1 and cch1) are shown in blue. With electrostimulation, they admit calcium into the cell. This calcium activates the the orange protein phosphatase calcineurin which dephosphorylates the purple transcription factor Crz1p. The dephosphorylated version of Crz1p, Crz1, is rapidly transported into the nucleus where it acts on the purple and blue promoters, transcribing GFP and/or RFP. Ultimately, our system did not get as far as having two coexisting promoters, but that was the plan.]] | ||
| + | <br> | ||
=='''Aims'''== | =='''Aims'''== | ||
| - | *Show that voltage can be used to stimulate a transcriptional response in S. cerevisiae. | + | *Show that voltage can be used to stimulate a transcriptional response in ''S. cerevisiae''. |
| - | *Develop a library of voltage-inducible | + | *Develop a library of voltage-inducible elements with differing voltage response curves. |
*Determine the functional range of and optimized values for our system with respect to the following variables: | *Determine the functional range of and optimized values for our system with respect to the following variables: | ||
**Voltage applied | **Voltage applied | ||
**Duration of voltage application | **Duration of voltage application | ||
| - | **Presence of | + | **Presence or absence of vesicular calcium pumps, yeast’s natural mode of intracellular calcium control |
*Develop an effective experimental apparatus to apply voltage and measure response. | *Develop an effective experimental apparatus to apply voltage and measure response. | ||
| + | |||
=='''Methods'''== | =='''Methods'''== | ||
===Visualizing the Crz1 Transcription Factor=== | ===Visualizing the Crz1 Transcription Factor=== | ||
| - | We | + | We wanted to visualize the behavior of Crz1 under electrostimulation, and confirm localization to the nucleus upon stimulation. The cells were induced, then mounted glass plate where they were imaged with a laser-scanning confocal microscope. Crz1-GFP was observed to move in and out of the nucleus just 5 minutes after a voltage stimulus. In a given image, some cells displayed Crz1-GFP densely packed in the nucleus while other cells displayed Crz1-GFP in the cytoplasm, but excluded from the nucleus. Here we see a confirmation of the oscillating behavior described by Cai ''et al''.<br> |
<html> | <html> | ||
<object width="640" height="385"><param name="movie" value="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="640" height="385"></embed></object> | <object width="640" height="385"><param name="movie" value="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HivMIxt6cAA?fs=1&hl=en_US" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="640" height="385"></embed></object> | ||
</html><br> | </html><br> | ||
| - | In | + | In the above video we can see the oscillatory behavior of Crz1-GFP after electrostimulation. Note that Crz1-GFP's oscillating movement into and out of the nucleus is not in phase throughout the cell population. Only a few cells actually exhibit a change in Crz1 localization during the course of the video. |
===Optimizing Parameters for the CDRE from the FKS2 Promoter=== | ===Optimizing Parameters for the CDRE from the FKS2 Promoter=== | ||
| - | We | + | We wanted to find an optimal voltage amplitude and electrostimulation duration for yeast containing the CDRE-RFP plasmid. Shocking at 10V caused widespread cell death, so it was necessary to find a balance between transcription and cellular damage. CDRE-RFP were grown and passed into 96 well plate. The electroporator was used to shock the cells with voltages from 2-10V for 0-80 seconds. We found that optimal transcription occurs when electrostimulation voltage is 8V. We also needed at least 40 seconds of electrostimulation to see expression. |
{| border="1" cellpadding="5" cellspacing="0" style="background: #ffffff;" | {| border="1" cellpadding="5" cellspacing="0" style="background: #ffffff;" | ||
| Line 80: | Line 86: | ||
|} | |} | ||
| - | + | Cells were analyzed using the above described confocal microscopy. As such, our data are purely quantitative. | |
| - | + | ||
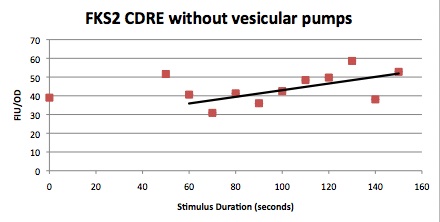
| - | [[Image:FSK2-CDRE.jpeg]] | + | ===Qualitative Characterization of the FKS2 CDRE=== |
| + | {| | ||
| + | |[[Image:FSK2-CDRE.jpeg|left|frame]] | ||
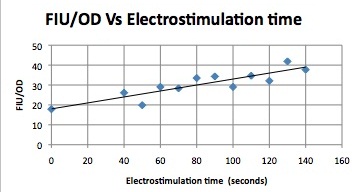
| + | ||Having decided on an optimal voltage, we then wanted to optimize the duration of electrostimulation. Additionally, these experiments provided the opportunity to compare yeast with and without functional PMC1 and VCX1 (vesicular calcium pumps). The data support a linear increase in expression of our RFP reporter with increased stimulation time. We also observe much higher expression in cells with vesicular pumps than in those without. | ||
| - | + | The graph to the left shows a increasing, linear relationship between electrostimulation time and expression of the RFP reporter. | |
| - | + | |} | |
| - | + | {| | |
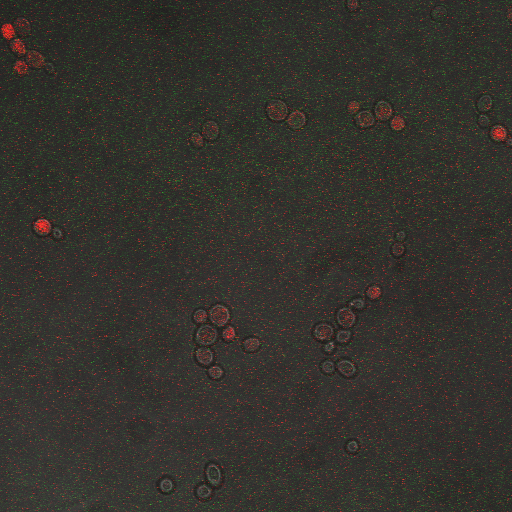
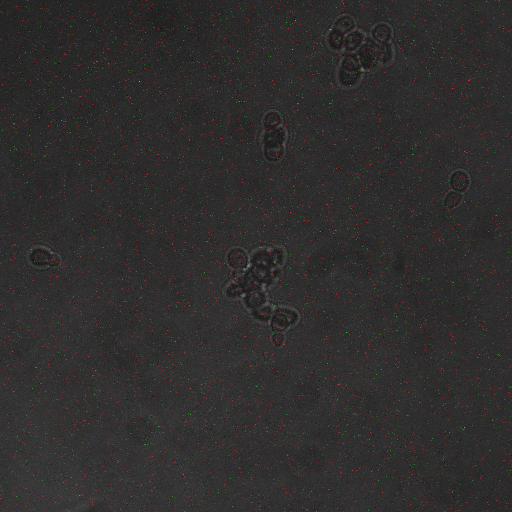
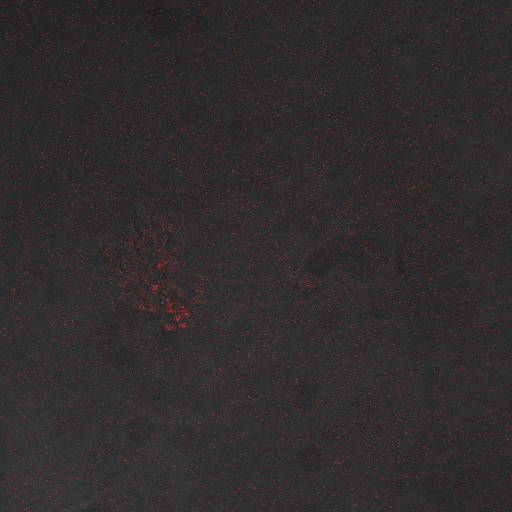
| + | |[[Image:Hone_in_0_Sec_KI.jpg|200px|thumb|FKS2 CDRE with vesicular pumps, 0 seconds electrostimulation]] | ||
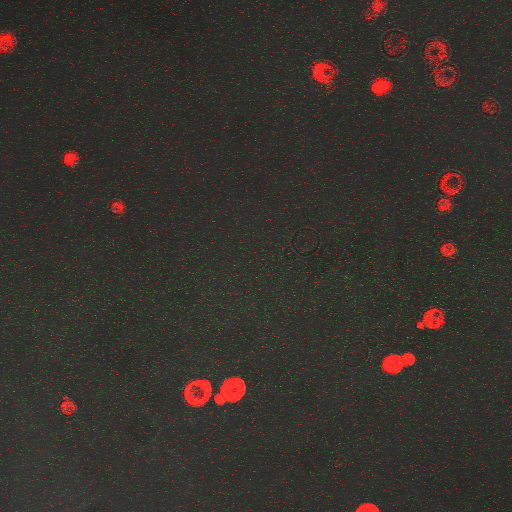
| + | |[[Image:Hone_in_90_Sec_KI.jpg|200px|thumb|FKS2 CDRE with vesicular pumps, 90 seconds electrostimulation]] | ||
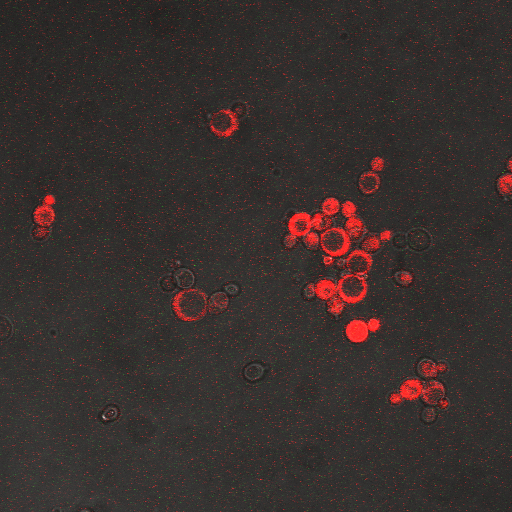
| + | |[[Image:Hone_in_110_Sec_KI.jpg|200px|thumb|FKS2 CDRE with vesicular pumps, 110 seconds electrostimulation]] | ||
| + | |} | ||
| + | In the images above we observe some constitutive expression of RFP in the FKS2 CDRE strain with the vesicular pumps. However, there is a linear increase in expression of the RFP reporter with increase in electrostimulation time up to 110 seconds. | ||
| + | <br> | ||
| + | {| | ||
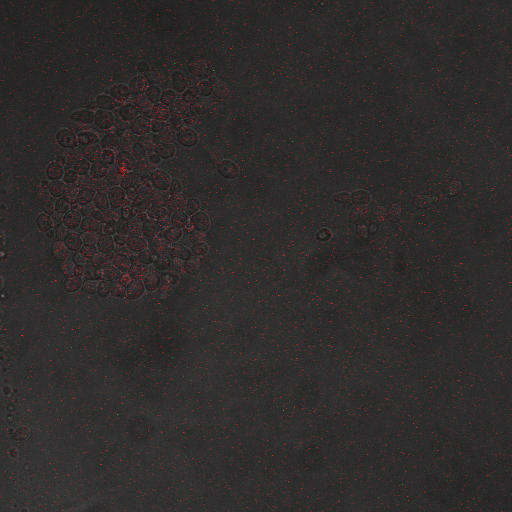
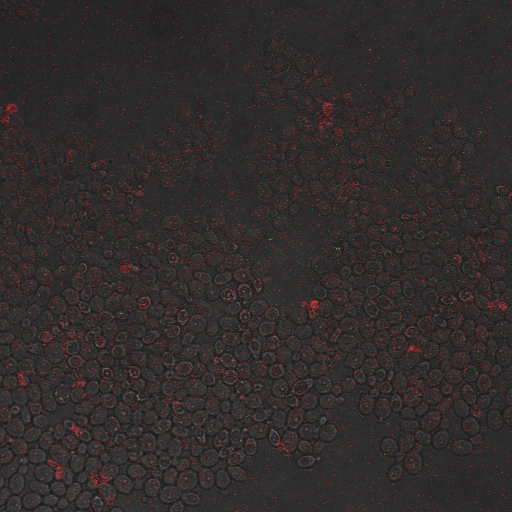
| + | |[[Image:Hone_in_80_Sec_KO.jpg|200px|thumb|FKS2 CDRE without vesicular pumps, 80 seconds electrostimulation]] | ||
| + | |[[Image:Hone_in_90_Sec_KO.jpg|200px|thumb|FKS2 CDRE without vesicular pumps, 90 seconds electrostimulation]] | ||
| + | |[[Image:Hone_in_110_Sec_KO.jpg|200px|thumb|FKS2 CDRE without vesicular pumps, 110 seconds electrostimulation]] | ||
| + | |[[Image:Hone_in_130_Sec_KO.jpg|200px|thumb|FKS2 CDRE without vesicular pumps, 130 seconds shocking]] | ||
| + | |} | ||
| + | Again, there is a linear relationship between stimulation duration and RFP reporter expression. However, in this case, the cells lacking vesicular pumps expressed less than those with vesicular pumps, and were less likely to be viable under electrostimulatory conditions. | ||
| - | + | ===Quantitative Characterization of FKS2 CDRE and PMC1 CDRE in Vacuole Positive and Negative Yeast=== | |
| - | + | At high time points, we observed that the unstimulated control often expressed as much as experimental samples. We postulated that this was a result of widespread cell death. While we normalized against OD, this can only account for cell density, not cell viability. We also noted that the data point at 40s (and later in one case) is consistently higher than many of the data with longer electrostimulation duration. We believe that this is the same effect. If this is so, it implies that the cell death that we observe is a steplike function of duration. | |
| - | + | ||
| - | [[Image: | + | In this experiment, cells with the FKS2 CDRE and the PMC1 CDRE were grown and passed into two rows of a 96 well plate and shocked at 8V with an exposure time of 0-80 seconds to quantitate the relationship between electrostimulation time and expression of a fluorescent protein. We again investigated the effect of knocking out the PMC1 and VCX1 vesicular calcium pumps. The data for both pump positive and pump negative cells are shown below.<br> |
| + | {| | ||
| + | |[[Image:fsk2 +.jpg|500px|thumb|This plot shows the fluorescence intensity normalized by OD 8 hours after induction by electrostimulation in cells positive for vesicular calcium pumps and using the FKS2 UAS in the reporter's promoter]] | ||
| + | |[[Image:fsk2 -.jpg|500px|thumb|This plot shows the fluorescence intensity normalized by OD 8 hours after induction by electrostimulation in cells negative for vesicular calcium pumps and using the FKS2 UAS in the reporter's promoter]] | ||
| + | |} | ||
| - | + | {| | |
| - | + | |[[Image:pmc1 +.jpg|550px|thumb|This plot shows the fluorescence intensity normalized by OD 8 hours after induction by electrostimulation in cells positive for vesicular calcium pumps and using the PMC1 UAS in the reporter's promoter]] | |
| - | + | |[[Image:Pmc1 final.jpg|550px|thumb|This plot shows the fluorescence intensity normalized by OD 8 hours after induction by electrostimulation in cells negative for vesicular calcium pumps and using the PMC1 UAS in the reporter's promoter]] | |
| - | + | |} | |
=='''Future Research Plans'''== | =='''Future Research Plans'''== | ||
| - | + | * Finish categorizing the synthetic CDREs using our high throughput electrostimulation device. | |
| + | * Apply our optimized high throughput experimentation technique to a library of synthetic CDRE sequences we have developed to characterize their voltage activation domains. We hope this library might allow others to have a more fine-grained control over the voltage response in their cells. | ||
| + | * Develop a genetic switch utilizing the repressible operator already present on FKS2. | ||
| + | * Develop an iGEM-compatible plasmid for yeast with a yeast-optimized origin of replication and markers. A shuttle vector would also be useful, enabling DNA to be stored or copied in bacteria and quickly transfered for testing in yeast. Our project would not be possible in a prokaryotic chassis. We feel that iGEM's apparent focus on ''E. coli'' may hinder the development of parts and systems that require eukaryotic organisms. | ||
==References== | ==References== | ||
| + | *Cyert MS. Calcineurin signaling in ''Saccharomyces cerevisiae'': how yeast go crazy in response to stress. Biochem Biophys Res Commun (2003) | ||
*Long Cai, Chiraj K. Dalal & Michael B. Elowitz. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature (2008). | *Long Cai, Chiraj K. Dalal & Michael B. Elowitz. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature (2008). | ||
| + | *Stathopoulos-Gerontides, ''et al''. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes & Development (1999). | ||
| + | *Yoshimoto ''et al''. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in ''Saccharomyces cerevisiae''. J Biol. Chem. (2002). | ||
Latest revision as of 03:53, 28 October 2010
Contents |
Abstract
If the goal of iGEM and the Parts Registry is to take the messy world of genetic engineering and transform it into something like the standardized world of electrical engineering, it may be useful if electronic systems could directly interface with biological systems. Past iGEM projects have used chemical or optical stimuli to actuate transcriptional responses. Our project, however, seeks to add voltage sensitivity to Saccharomyces cerevisiae. Baker’s yeast was chosen because in some sense yeast have a system that responds to voltage input. With a voltage stimulus one can open the voltage-gated calcium channels of yeast, causing calcium ions to rush into the cytoplasm. This causes calcineurin to dephosphorylate Crz1, which enters the nucleus and binds various promoters. Our group presents a library of characterized Crz1-sensitive binding sites of both naturally-occurring and synthetic varieties that can be integrated into promoters. Genes downstream of these promoters are thus voltage-regulated in media containing calcium.
Aims
- Show that voltage can be used to stimulate a transcriptional response in S. cerevisiae.
- Develop a library of voltage-inducible elements with differing voltage response curves.
- Determine the functional range of and optimized values for our system with respect to the following variables:
- Voltage applied
- Duration of voltage application
- Presence or absence of vesicular calcium pumps, yeast’s natural mode of intracellular calcium control
- Develop an effective experimental apparatus to apply voltage and measure response.
Methods
Visualizing the Crz1 Transcription Factor
We wanted to visualize the behavior of Crz1 under electrostimulation, and confirm localization to the nucleus upon stimulation. The cells were induced, then mounted glass plate where they were imaged with a laser-scanning confocal microscope. Crz1-GFP was observed to move in and out of the nucleus just 5 minutes after a voltage stimulus. In a given image, some cells displayed Crz1-GFP densely packed in the nucleus while other cells displayed Crz1-GFP in the cytoplasm, but excluded from the nucleus. Here we see a confirmation of the oscillating behavior described by Cai et al.
In the above video we can see the oscillatory behavior of Crz1-GFP after electrostimulation. Note that Crz1-GFP's oscillating movement into and out of the nucleus is not in phase throughout the cell population. Only a few cells actually exhibit a change in Crz1 localization during the course of the video.
Optimizing Parameters for the CDRE from the FKS2 Promoter
We wanted to find an optimal voltage amplitude and electrostimulation duration for yeast containing the CDRE-RFP plasmid. Shocking at 10V caused widespread cell death, so it was necessary to find a balance between transcription and cellular damage. CDRE-RFP were grown and passed into 96 well plate. The electroporator was used to shock the cells with voltages from 2-10V for 0-80 seconds. We found that optimal transcription occurs when electrostimulation voltage is 8V. We also needed at least 40 seconds of electrostimulation to see expression.
| 2s | 5s | 10s | 20s | 40s | 80s | |
|---|---|---|---|---|---|---|
| 10V | None | None | None | None | None | N/A |
| 8V | None | None | None | None | High | High |
| 6V | None | None | None | None | Moderate | High |
| 4V | None | None | None | None | Low | Low |
Cells were analyzed using the above described confocal microscopy. As such, our data are purely quantitative.
Qualitative Characterization of the FKS2 CDRE
| Having decided on an optimal voltage, we then wanted to optimize the duration of electrostimulation. Additionally, these experiments provided the opportunity to compare yeast with and without functional PMC1 and VCX1 (vesicular calcium pumps). The data support a linear increase in expression of our RFP reporter with increased stimulation time. We also observe much higher expression in cells with vesicular pumps than in those without.
The graph to the left shows a increasing, linear relationship between electrostimulation time and expression of the RFP reporter. |
In the images above we observe some constitutive expression of RFP in the FKS2 CDRE strain with the vesicular pumps. However, there is a linear increase in expression of the RFP reporter with increase in electrostimulation time up to 110 seconds.
Again, there is a linear relationship between stimulation duration and RFP reporter expression. However, in this case, the cells lacking vesicular pumps expressed less than those with vesicular pumps, and were less likely to be viable under electrostimulatory conditions.
Quantitative Characterization of FKS2 CDRE and PMC1 CDRE in Vacuole Positive and Negative Yeast
At high time points, we observed that the unstimulated control often expressed as much as experimental samples. We postulated that this was a result of widespread cell death. While we normalized against OD, this can only account for cell density, not cell viability. We also noted that the data point at 40s (and later in one case) is consistently higher than many of the data with longer electrostimulation duration. We believe that this is the same effect. If this is so, it implies that the cell death that we observe is a steplike function of duration.
In this experiment, cells with the FKS2 CDRE and the PMC1 CDRE were grown and passed into two rows of a 96 well plate and shocked at 8V with an exposure time of 0-80 seconds to quantitate the relationship between electrostimulation time and expression of a fluorescent protein. We again investigated the effect of knocking out the PMC1 and VCX1 vesicular calcium pumps. The data for both pump positive and pump negative cells are shown below.
Future Research Plans
- Finish categorizing the synthetic CDREs using our high throughput electrostimulation device.
- Apply our optimized high throughput experimentation technique to a library of synthetic CDRE sequences we have developed to characterize their voltage activation domains. We hope this library might allow others to have a more fine-grained control over the voltage response in their cells.
- Develop a genetic switch utilizing the repressible operator already present on FKS2.
- Develop an iGEM-compatible plasmid for yeast with a yeast-optimized origin of replication and markers. A shuttle vector would also be useful, enabling DNA to be stored or copied in bacteria and quickly transfered for testing in yeast. Our project would not be possible in a prokaryotic chassis. We feel that iGEM's apparent focus on E. coli may hinder the development of parts and systems that require eukaryotic organisms.
References
- Cyert MS. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun (2003)
- Long Cai, Chiraj K. Dalal & Michael B. Elowitz. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature (2008).
- Stathopoulos-Gerontides, et al. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes & Development (1999).
- Yoshimoto et al. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol. Chem. (2002).
 "
"