|
|
| (30 intermediate revisions not shown) |
| Line 86: |
Line 86: |
| | </html> | | </html> |
| | {|align="center" | | {|align="center" |
| | + | [[Image:Virginia_United_2.jpg|714 px|center|]] |
| | | | | | |
| - | |-
| + | == '''Quorum Sensing Amplifiers and a Co-design Approach for Information Processing''' == |
| - | |
| + | ==== Abstract ==== |
| - | ''' | + | Synthetic biology endeavors to create information processing systems modeled on digital electronics. The use of quorum sensing can help transform an inherently analog molecular signal into a binary response and simultaneously allow the tuning of input response thresholds and signal amplification. This project demonstrates these capabilities through experimentation and modeling. Another candidate for reapplying an electronic engineering technique is the codesign of hardware and software to implement a function. In synthetic biology, codesign might mean implementing a design spec in different expression control regimes and comparing their relative merits. Our work examines the codesign concept by constructing an AND gate in three different design domains. We explore the application of these ideas with an environmental sensor. A unique aspect of our project is the collaborative nature involving five institutions at three locations, which fostered a codesign-like approach using two distinct assembly techniques. |
| - | |[[Image:Virginia_United_2.jpg|550px|thumb|left]]
| + | |
| - | [[Image:vaunited.jpg|250px|thumb|right]]
| + | |
| - | |-
| + | ---- |
| - | |
| + | |
| - | |}
| + | ==== Quorum Sensing ==== |
| - | == '''An Engineering Approach to an Environmental Biosensor''' == | + | |
| | + | Normally, quorum sensing is a mechanism employed by cells in which signaling proteins are released and passed on to other cells in order to communicate certain information or to encourage other cells in the vicinity to act out certain functions. In our experiment, we wanted to utilize quorum sensing so that when the E. coli cells sensed that there was a input in the environment, it would not only produce some type of fluorescent response, but also initiate the quorum sensing mechanism so that the signal would be amplified and passed on to other cells that would then react and fluoresce as well. In that manner, instead of receiving a gradual response to the stimuli, we would actually receive a quicker, more binary response that would be easier to detect and measure. Thus in our experiment, we wanted to employ quorum sensing as an amplification mechanism that would provide us with binary output, making the system easier to manipulate into a quantitative response system. |
| | + | |
| | + | |
| | + | ---- |
| | | | |
| - | In engineering, it is recognized that there are various ways to successfully design a mechanism. Often, different designs are tested and compared given a set of metrics in order to assess the strengths and weaknesses of each option. This process is called co-design. Since synthetic biology is truly a multi-disciplinary field, it is important that we incorporate techniques that have been proven successful in other well-established disciplines. We decided to apply co-design while testing and evaluating the effectiveness of a multiple-compound biosensor detection process.
| + | ==== Co-Design ==== |
| | | | |
| | [[Image: Project800px-VA-720.jpg|720px|thumb|center|]] | | [[Image: Project800px-VA-720.jpg|720px|thumb|center|]] |
| | | | |
| - | In order to compare our designs, we are implementing logic on three different regulatory levels of the cell. One approach utilizes the operator sites of regulatory promoters, hybridizing two promoters’ operator sites into a single co-sensing promoter. In order for the hybrid promoter to initiate transcription, both target compounds that control the operator sites must be present. The hybrid promoters are attached to a single fluorescent protein, so detection of both target compounds can be measured via fluorescence.
| + | ===== Fluorescent Complementation ===== |
| | | | |
| - | Another method utilizes fluorescent protein complementation. When a target molecule is detected by a cell, it will transcribe a non-fluorescent half of a protein. When another target molecule is detected, the other half of the fluorescent protein is transcribed. Upon translation, the halves will bond together and fluoresce, reporting the presence of the two target compounds.
| + | Magliery et. al (2004) developed a method for detecting protein binding partners by fusing non-fluorescent fragments of GFP to the two peptides of interest. If the attached peptides sufficiently attract one another, the GFP |
| | + | fragments will be irreversibly reassemble to form a fluorescing GFP protein. We build on their work by demonstrating how GFP fragment reassembly ("fluorescent complementation") can be used to create AND logic. |
| | | | |
| - | The final approach allows each of the sensory reporters to express a fluorescent protein in the presence of its target compound. If multiple target compounds are detected by a culture, fluorescence spectroscopy is used to separate out the wavelengths of each fluorescent protein, which then determines what compounds are present in the system. In all three designs we are amplifying the signal that each E. coli cell emits once it is exposed to the target compound with a quorum sensing system. Each cell releases a signal when exposed to the target compound, which is then recognized by neighboring cells. A fluorescent protein is attached to the promoter that recognizes the signal, establishing a more rapid, binary-like response time in the system.
| + | ===== Hybrid Promoters ===== |
| | | | |
| - | We decided to evaluate our co-designs by developing a biosensor that detects the presence of mercury, copper, and arsenic in aquatic environments. In small doses each metal may not individually be toxic to fish, but a combination of the metals, even if individually each metal is at a negligible volume, may still be hazardous to both the fish in such environments and the people that later consume those fish.
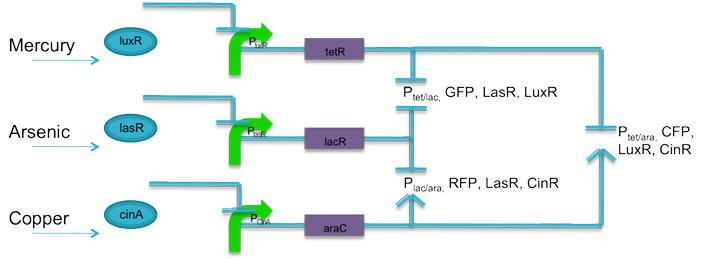
| + | The advancement of synthetic biology has opened the doors to almost limitless transcriptional control, given the particular biological species cooperates. The aim of the hybrid promoter circuit is to give one fluorescent output based on any two inputs. To do this, logic AND gates are designed using promoters and protein operator sites. |
| | | | |
| - | As a final product, we hope that the system can be utilized not just for testing aquatic toxicity levels, but can serve as a basis for a synthetic machinery that has interchangeable inputs and outputs so it can be used for other applications such as biosecurity. Additionally, with an interchangeable response, it is possible to not only implement a biosensor within this system, but in the future, bioremediation machinery could be applied as an output as well.
| + | [[Image:HybridPromoterSystem-720.png|720px|thumb|center]] |
| | | | |
| - | == Project Details==
| + | Using literature, current biobricks, and promoters J23106 and J23110 as base promoters, two sets of promoters were designed to respond to tetracycline and lactose, tetracycline and arabinose, and lactose and arabinose. The promoters are predicted to have similar transcription rates as their base promoters. According to literature, a promoter with an activation site that is not activated is not entirely inactive. RNA polymerase can still bind, only the interaction is weaker. Promoters J23106 and J23110 were chose because they are of relatively moderate strength. It is predicted that until arabinose is present in the system, the relative strength of these promoters will be fairly weak. When coupled with weak ribosomal binding sites, the rate at which proteins are translated should be negligible, preventing false positives. Also, following each fluorescent protein, is the respective quorum sensing protein to keep the system active. |
| | | | |
| - | ===== Abstract =====
| + | [[Image:HybridAndGate-720.png|720px|thumb|center]] |
| | | | |
| - | Synthetic biology endeavors to create information processing systems modeled on digital electronics. The use of quorum sensing can help transform an inherently analog molecular signal into a binary response and simultaneously allow the tuning of input response thresholds and signal amplification. This project demonstrates these capabilities through experimentation and modeling. Another candidate for reapplying an electronic engineering technique is the codesign of hardware and software to implement a function. In synthetic biology, codesign might mean implementing a design spec in different expression control regimes and comparing their relative merits. Our work examines the codesign concept by constructing an AND gate in three different design domains. We explore the application of these ideas with an environmental sensor. A unique aspect of our project is the collaborative nature involving five institutions at three locations, which fostered a codesign-like approach using two distinct assembly techniques.
| + | ---- |
| | | | |
| - | ===== Regional Team ===== | + | ===== Optimization of Fluorescent Proteins ===== |
| | | | |
| - | The Virginia United Team was created as a collaborative effort between five schools, University of Virginia, Virginia Commonwealth University, Virginia Polytechnic Institute and State University (Virginia Tech), Bluefield State College, and Virginia State University. We divided up and worked on separate parts of the project in three different locations in the state and maintained communication through constant emails, Skype calls, and face to face regional meetings. Collaborating on a project of this magnitude with this many people involved was in itself an experimental endeavor that challenged us to develop a project and maintain strong communication and organizational skills that are vital to the interdisciplinary component of synthetic biology. Not only did each individual have something unique to offer to the team, but so did each individual university based on their access to resources that other schools might have lacked. Perhaps one of the most vital components of this project is what we learned from working together to help develop a project that we would not have been able to accomplish individually. Having gained a better understanding of how research can be conducted in a huge group setting has provided us with the experience that will really become instrumental in our future in research and in particular synthetic biology as the need for collaborative projects between schools, states, and even countries becomes a normal part of the research world.
| + | Optimization of protein expression is an essential skill in synthetic biology; the ability to tweak protein expression to the levels one desires eases the process of designing precise biological systems. Fluorescent proteins function as appropriate applications in optimization experiments because fluorescence expression levels can easily be measured by a spectrofluorometer or plate reader. |
| | | | |
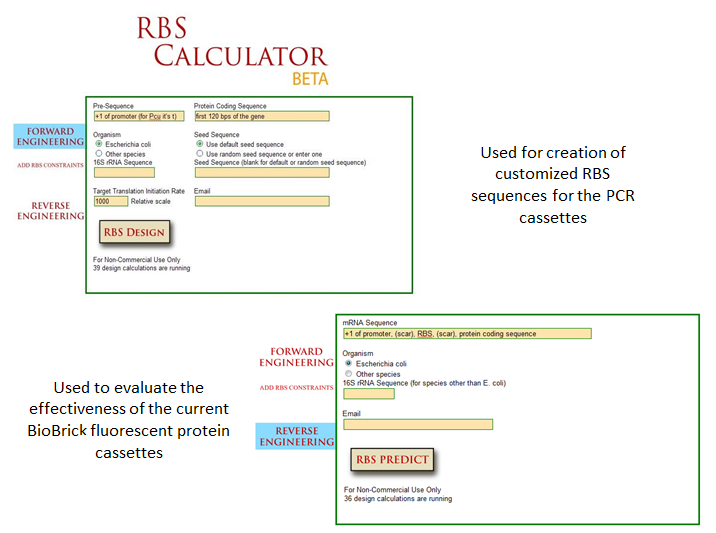
| - | ===== Collaborations with Other Teams =====
| + | [[Image:RBS Calculator.png|720px|thumb|center]] |
| | | | |
| - | ''We helped the software team create a program that can analyze a
| + | One method of optimization that has recently been explored is the inherent differences in efficiency of gene-specific ribosome binding sites (Voigt et al., 2009). The majority of the time when designing a construct in iGEM, a ‘default’ RBS is used by including just the basic Shine-Dalgarno sequence found in all prokaryotic ribosome binding sites. What many do not realize is that when utilizing the Shine-Delgarno conserved sequence in different gene constructs, the efficiency of the expression for that construct depends on the gene. Utilizing the RBS calculator developed by Voigt et al., one can test the actual optimization rates of the ribosome binding sites by measuring fluorescence output of the final constructs. This allows more control in the system, instituting more flexibility in the precise design of one’s constructs. |
| - | construct's DNA sequence and design oligo primers that can be used for USER | + | |
| - | fusion. Our team helped by explaining how USER fusion works, explaining
| + | |
| - | optimal length of primers, and explaining optimal melting temperatures for
| + | |
| - | the primers. USER fusion is a method for splicing together long strands of | + | |
| - | DNA and is convenient for construct assembly.''
| + | |
| | | | |
| - | ===== Overview =====
| + | ---- |
| | | | |
| - | As the field of biomedical engineering grows, so too does the complexity of genetically engineered systems. Fabricated metabolic pathways are becoming increasingly intricate and expansive: how is one to know if the approach used to synthesize the pathway is necessarily the best approach? Our group, composed of five institutions, strives to show that co-design, a common engineering practice used to evaluate relative efficiencies, can be implemented to select the best design for a system. Using a biosensor for an example of carrying out co-design, we constructed three systems utilizing different mechanisms that can detect inputs and give an output correlating to which input or inputs it senses. To maintain consistency, our systems are designed to use quorum sensing to amplify signal transductionupon input detection. This essentially acts as an on/off switch, allowing the system to be engineered to detect a certain input threshold and giving a binary response.
| + | ==== References ==== |
| | | | |
| - | ===== Quorum Sensing =====
| + | Ball DA, Lux MW, Graef RR, Peterson MW, Valent JD, Dileo J, Peccoud J (2010) Co-design in Synthetic Biology: A System-Level Analysis of the Development of an Environmental Sensing Device. Biocomputing 2010: 385-396. |
| | | | |
| - | ''Normally, quorum sensing is a mechanism employed by bacterial cells in which signaling proteins are released and passed on to other cells so communicate certain information or to encourage cells in the vicinity to act out certain functions. In our experiment, we wanted to utilize quorum sensing so that when the E. coli cells sensed that there was a metal in the environment, it would not only produce some type of response such as fluorescing, but also initiate the quorum sensing mechanism so that the signal would be amplified and passed on to other cells that would then react and flouress as well. In that manner, instead of receiving a gradual response to the stimuli, we would actually receive a faster more binary response that would be easier to detect and measure. Thus in our experiment, we wanted to employ quorum sensing as an amplification mechanism that would provide us with binary output which would be easier to manipulate and incorporate into a response system.''
| + | Balagadde FK, Song H, Ozaki J, Collins C, Barnet M, Arnold FH, Quake SR, You L (2008) A Synthetic Escherichia coli Predator-Prey Ecosystem. Molecular Systems Biology 4: 187 |
| | | | |
| - | ==== Fluorescent Complementation ====
| + | Collins CH, Arnold FH, Leadbetter JR (2005). Directed Evolution of Vibrio fischeri LuxR for Increased Sensitivity to a Broad Spectrum of Acyl-homoserine Lactones. Molecular Microbiology 55: 712-723. |
| | | | |
| - | Magliery et. al (2004) developed a method for detecting protein binding partners by fusing non-fluorescent fragments of GFP to the two peptides of interest. If the attached peptides sufficiently attract one another, the GFP
| + | Collins CH, Leadbetter JR, Arnold FH (2006). Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nature Biotechnology 24: 708-712. |
| - | fragments will be irreversibly reassemble to form a fluorescing GFP protein. We build on their work by demonstrating how GFP fragment reassembly ("fluorescent complementation") can be used to create AND logic.
| + | |
| | | | |
| - | Magliery et al. Detecting Protein- Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism. J. Am.
| + | Czar CJ, Anderson C, Bader JS, Peccoud J (2009) Gene Synthesis Demystified. Trends in Biotechnology 10: 63-72. |
| - | Chem. Soc (2005) vol. 127 (1) pp. 146-157
| + | |
| | | | |
| - | ====Hybrid Promoters====
| + | Fuqua C, Winans S, Greenberg EP (1996). Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol 50: 727-751. |
| | | | |
| - | The advancement of synthetic biology has opened the doors to almost limitless transcriptional control, given the particular biological species cooperates. The aim of the hybrid promoter circuit is to give one fluorescent output based on any two inputs. To do this, logic AND gates are designed using promoters and protein operator sites.
| + | Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339-342. |
| | | | |
| - | [[Image:HybridPromoterSystem-720.png|720px|thumb|center]]
| + | Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA (2007). USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Research 35: E55. |
| | | | |
| - | Using literature, current biobricks, and promoters J23106 and J23110 as base promoters, two sets of promoters were designed to respond to tetracycline and lactose, tetracycline and arabinose, and lactose and arabinose. The promoters are predicted to have similar transcription rates as their base promoters. According to literature, a promoter with an activation site that is not activated is not entirely inactive. RNA polymerase can still bind, only the interaction is weaker. Promoters J23106 and J23110 were chose because they are of relatively moderate strength. It is predicted that until arabinose is present in the system, the relative strength of these promoters will be fairly weak. When coupled with weak ribosomal binding sites, the rate at which proteins are translated should be negligible, preventing false positives. Also, following each fluorescent protein, is the respective quorum sensing protein to keep the system active.
| + | Magliery et al. Detecting Protein- Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism. J. Am. Chem. Soc (2005) vol. 127 (1) pp. 146-157 |
| | | | |
| - | [[Image:HybridAndGate-720.png|720px|thumb|center]]
| + | Mukerji S, Oudenaarden A (2009) Synthetic Biology: understanding biological design from synthetic circuits. Nature Reviews Genetics 10: 859-871. |
| | + | |
| | + | Nasser W, Bouillant ML, Salmond G, Reverchon S (1998). Characterization of the Erwinia chrysnthemi expI – expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Molecular Microbiology 6: 1391-1405. |
| | + | |
| | + | Salis, H., Mirsky, E.A. and Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression, Nat. Biotechnol. 27, 946 - 950 (2009) |
 "
"