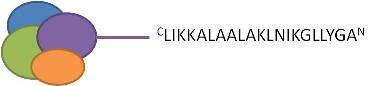Team:Stockholm/30 June 2010
From 2010.igem.org
(Difference between revisions)
(New page: {{Stockholm/Top2}} = Morning meeting = We discussed the project proceedings. * It was decided that already amplified genes (cloned in pEX vector) should be transferred to [http://partsre...) |
m |
||
| (5 intermediate revisions not shown) | |||
| Line 22: | Line 22: | ||
= Andreas = | = Andreas = | ||
| - | == Expression of SOD and yCCS from pEX expression vector == | + | === Expression of SOD and yCCS from pEX expression vector === |
''Continued from 29/6'' | ''Continued from 29/6'' | ||
| - | We realized that Top10 and DH5alpha are not suitable for IPTG-induced protein expression. SOD and yCCS expression was therefore not proceeded from ON cultures. '''Glycerol stocks''' were prepared from ON culture and '''pEX | + | We realized that Top10 and DH5alpha are not suitable for IPTG-induced protein expression. SOD and yCCS expression was therefore not proceeded from ON cultures. '''Glycerol stocks''' were prepared from ON culture and '''pEX.SOD''' and '''pEX.yCCS plasmids''' were prepared. |
| - | == Preparation of competent BL21(DE3) cells == | + | '''DNA concentrations''' |
| - | * | + | {|border="1" cellspacing="0" cellpadding="2" |
| + | |'''Sample''' | ||
| + | |'''DNA conc (ng/ul)''' | ||
| + | |'''A(260)/A(280)''' | ||
| + | |- | ||
| + | |pEX.SOD | ||
| + | |18.15 | ||
| + | |1.98 | ||
| + | |- | ||
| + | |pEX.yCCS | ||
| + | |20.55 | ||
| + | |1.97 | ||
| + | |} | ||
| + | |||
| + | === Preparation of competent BL21(DE3) cells === | ||
| + | * 5 ml LB was inoculated with BL21(DE3) cells and grown ON in 37°C, 250 rpm. | ||
| + | |||
| + | |||
| + | |||
| + | = Mimmi = | ||
| + | |||
| + | === CPP's === | ||
| + | |||
| + | ==== designing primers for assembly standard 25 ==== | ||
| + | |||
| + | *No start or stop codon | ||
| + | |||
| + | *From the pSB1AsF: | ||
| + | |||
| + | [[Image:primer-design.jpg]] | ||
| + | |||
| + | *Only the length can be ajusted, too short with 16 bases? | ||
| + | |||
| + | ===== Transportan10 ===== | ||
| + | |||
| + | *Probably good to bind in the C-terminal end (not tested): | ||
| + | |||
| + | [[Image:Transportan10.jpg]] | ||
| + | |||
| + | :To sit in the N-terminal end of the protein of interest: | ||
| + | ::*Sequence the right way, use the n-part prefix | ||
| + | |||
| + | G . . AATTC--------------114---------------CTGCA . . G | ||
| + | |||
| + | CTTAA . . G--------------106---------------G . . ACGTC | ||
| + | |||
| + | :To sit in the C-terminal end of the protein of interest: | ||
| + | ::*turn the aa sequence around -> the DNA sequence around, use the normal prefix | ||
| + | |||
| + | |||
| + | ===== LMWP ===== | ||
| + | |||
| + | *Binding in the N-terminal (as done in the study): | ||
| + | |||
| + | [[Image:LMWP.jpg]] | ||
| + | |||
| + | :To sit in the N-terminal end of the protein of interest: | ||
| + | ::*turn the aa sequence around -> the DNA sequence around, use the n-part prefix | ||
| + | |||
| + | :To sit in the C-terminal end of the protein of interest: | ||
| + | ::*Sequence the right way, use the normal prefix | ||
| + | |||
| + | |||
| + | ===== TAT (dowdy's) ===== | ||
| + | |||
| + | *Binding in both terminals (neither is done in the study): | ||
| + | |||
| + | [[Image:TAT.jpg]] | ||
| + | |||
| + | =Johan= | ||
| + | |||
| + | * Miniprep | ||
| + | |||
| + | Promega plasmid yield | ||
| + | |||
| + | 2 tubes bFGF, 3 ml from each (maximum for that protocol) | ||
| + | |||
| + | Abs: 20 ng/µl & 70 ng/µl | ||
| + | |||
| + | * Miniprep | ||
| + | |||
| + | - Rest of bFGF (12 ml in total) | ||
| + | - 12 ml * 5 pEX | ||
| + | |||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 18:38, 27 October 2010
Contents |
Morning meeting
We discussed the project proceedings.
- It was decided that already amplified genes (cloned in pEX vector) should be transferred to [http://partsregistry.org/Part:pSB1C3 pSB1C3] vector.
- IgG protease
- Superoxidase dismutase (SOD)
- yCCS
- bFGF
- Primers for not-yet amplified genes should be redesigned to include the complete Assembly standard 25 prefix and suffix.
- CPP
- Transportan 10
- LMWP
- TAT
- MITF
- Protein A Z-domain
- Tyrosinase
- Vitamin B9 genes
- CPP
- BL21(DE3) was chosen as our strain for IPTG-induced protein expression from pEX vector.
Andreas
Expression of SOD and yCCS from pEX expression vector
Continued from 29/6
We realized that Top10 and DH5alpha are not suitable for IPTG-induced protein expression. SOD and yCCS expression was therefore not proceeded from ON cultures. Glycerol stocks were prepared from ON culture and pEX.SOD and pEX.yCCS plasmids were prepared.
DNA concentrations
| Sample | DNA conc (ng/ul) | A(260)/A(280) |
| pEX.SOD | 18.15 | 1.98 |
| pEX.yCCS | 20.55 | 1.97 |
Preparation of competent BL21(DE3) cells
- 5 ml LB was inoculated with BL21(DE3) cells and grown ON in 37°C, 250 rpm.
Mimmi
CPP's
designing primers for assembly standard 25
- No start or stop codon
- From the pSB1AsF:
- Only the length can be ajusted, too short with 16 bases?
Transportan10
- Probably good to bind in the C-terminal end (not tested):
- To sit in the N-terminal end of the protein of interest:
- Sequence the right way, use the n-part prefix
G . . AATTC--------------114---------------CTGCA . . G
CTTAA . . G--------------106---------------G . . ACGTC
- To sit in the C-terminal end of the protein of interest:
- turn the aa sequence around -> the DNA sequence around, use the normal prefix
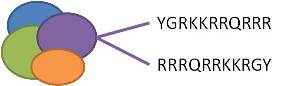
LMWP
- Binding in the N-terminal (as done in the study):
- To sit in the N-terminal end of the protein of interest:
- turn the aa sequence around -> the DNA sequence around, use the n-part prefix
- To sit in the C-terminal end of the protein of interest:
- Sequence the right way, use the normal prefix
TAT (dowdy's)
- Binding in both terminals (neither is done in the study):
Johan
- Miniprep
Promega plasmid yield
2 tubes bFGF, 3 ml from each (maximum for that protocol)
Abs: 20 ng/µl & 70 ng/µl
- Miniprep
- Rest of bFGF (12 ml in total) - 12 ml * 5 pEX
 |

|
 |

|
 |

|

|

|
 "
"