Team:Stockholm/15 October 2010
From 2010.igem.org
NinaSchiller (Talk | contribs) (→Andreas) |
|||
| (One intermediate revision not shown) | |||
| Line 168: | Line 168: | ||
====Results==== | ====Results==== | ||
Results indicate no significant difference in growth between different cultures. Slightly lower curve for the nTra10-expressing clone, which is somewhat interesting, since a similar progress was seen in the canceled experiment from 14/10. At least one more experiment will be run next week, using new BL21 clones; this will show whether this is just a coincidence or not. | Results indicate no significant difference in growth between different cultures. Slightly lower curve for the nTra10-expressing clone, which is somewhat interesting, since a similar progress was seen in the canceled experiment from 14/10. At least one more experiment will be run next week, using new BL21 clones; this will show whether this is just a coincidence or not. | ||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === clone non-CPP operon === | ||
| + | |||
| + | ==== colony PCR ==== | ||
| + | |||
| + | {| | ||
| + | ! mix | ||
| + | | | ||
| + | | x16 | ||
| + | | rowspan="5" width="150" | | ||
| + | ! primers | ||
| + | | rowspan="5" width="150" | | ||
| + | ! samples | ||
| + | |- | ||
| + | | mastermix | ||
| + | | 24.5 | ||
| + | | | ||
| + | | pSB_VF2 | ||
| + | | pSB1C3.SOD.RBS.yCCS 1-5 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 0.5 | ||
| + | | | ||
| + | | pSB_VR | ||
| + | | pSB1C3.SOD.his.RBS.yCCS 1-5 | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 25µl | ||
| + | | | ||
| + | | rowspan="2" | | ||
| + | | pSB1C3.his.SOD.RBS.yCCS 1-5 | ||
| + | |- | ||
| + | | colspan="3" | | ||
| + | | positive control (pSB1C3.LMWP.SOD.his) | ||
| + | |} | ||
| + | |||
| + | ==== Gel ==== | ||
| + | |||
| + | [[Image:2010-10-15 pC.S yC.jpg|200px|thumb|left|]] | ||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | | 12 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | pC.S_yC 1 | ||
| + | | 13 | ||
| + | | pC.hS_yC 1 | ||
| + | |- | ||
| + | | 3 | ||
| + | | pC.S_yC 2 | ||
| + | | 14 | ||
| + | | pC.hS_yC 2 | ||
| + | |- | ||
| + | | 4 | ||
| + | | pC.S_yC 3 | ||
| + | | 15 | ||
| + | | pC.hS_yC 3 | ||
| + | |- | ||
| + | | 5 | ||
| + | | pC.S_yC 4 | ||
| + | | 16 | ||
| + | | pC.hS_yC 4 | ||
| + | |- | ||
| + | | 6 | ||
| + | | pC.S_yC 5 | ||
| + | | 17 | ||
| + | | pC.hS_yC 5 | ||
| + | |- | ||
| + | | 7 | ||
| + | | pC.Sh_yC 1 | ||
| + | | 18 | ||
| + | | pC.L.hS, control | ||
| + | |- | ||
| + | | 8 | ||
| + | | pC.Sh_yC 2 | ||
| + | | rowspan="4" | | ||
| + | | rowspan="4" | | ||
| + | |- | ||
| + | | 9 | ||
| + | | pC.Sh_yC 3 | ||
| + | |- | ||
| + | | 10 | ||
| + | | pC.Sh_yC 4 | ||
| + | |- | ||
| + | | 11 | ||
| + | | pC.Sh_yC 5 | ||
| + | |} | ||
| + | |||
| + | |||
| + | ==== ON culture ==== | ||
| + | |||
| + | 3ml LB Cm<sub>25</sub> + toothpick from colony | ||
| + | |||
| + | SOD.RBS.yCCS 5 | ||
| + | SOD.his.RBS.yCCS 2 | ||
| + | his.SOD.RBA.yCCS 3 | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 11:10, 26 October 2010
Contents |
Nina
Continuation of protein purification
Washing
I washed the column with 10 X volumes of wash buffer.
The Ni-resin (in the form of pellet) is 10 ml, therefore 10 * 10ml Ni-NTA = 100 ml wash buffer.
The imidazole is 2 M.
2M * Volume = 100 * 30 *10^-3 M Volume imidaziole that needs to be added in the wash 100 ml is 1.5 ml
Washing:
Elution
I eluted the column with 5 X volumes of elution buffer.
The Ni-resin (in the form of pellet) is 10 ml, therefore 5 * 10ml Ni-NTA = 50 ml elution buffer.
- 20 ml 40 min on shake 4°C
- 10 ml 30 min on shake 4°C
- 10 ml 30 min on shake 4°C
- 10 ml 30 min on shake 4°C
I saved these samples in the cold room 4°C for tomorrow.
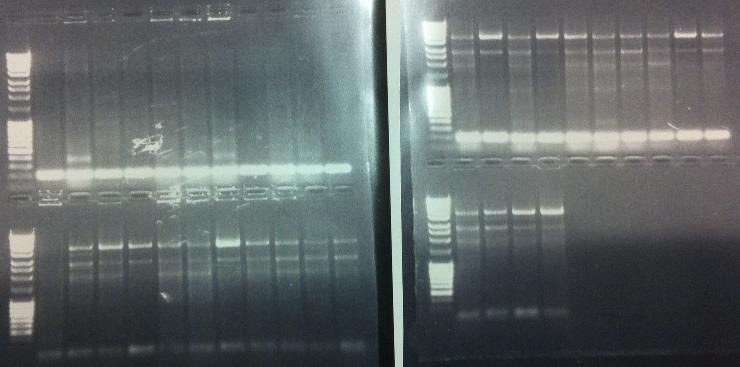
Agarose gel verification
I ran the PCR products on an 1 % agarose gel 80 V to check if I had any inserts.
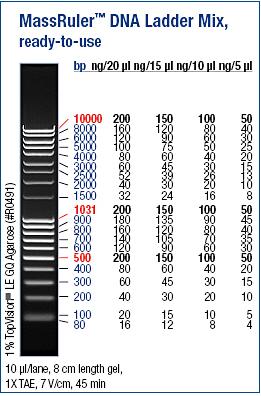
Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp
Arrangement on gel:
Gels:
Unfortunately non of the bands show that there has been an insert, it might be that the transformation into BL21 cells was not a good idea since they are not cloning cells, but we are short on time and I tried to make a short cut. It might have failed this time. I will run a new screen with 16 colonies/dish, it that won't work then this cloning has not worked out well.
Andreas
Growth curve assay
Setup
Inoculated new flasks from ON cultures set 14/10:
- 250 ml E-flasks
- 25 ml LB + Amp 100 + 1 mM IPTG
- 250 μl ON culture
- BL21 pEX.SOD⋅His
- BL21 pEX.nTra10⋅SOD⋅His
- BL21 pEX.nTAT⋅SOD⋅His
- BL21 pEX.nLMWP⋅SOD⋅His
- 37 °C, 225 rpm
OD590 measurements made every 30 min with appropriate dilution in fresh LB.
OD590 measurements
| 0 min | 30 min | 60 min | 90 min | 120 min | 150 min | 180 min | 210 min | 240 min | 270 min | 300 min | 330 min | 360 min | 390 min | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pEX.SOD⋅His | 0.035 | 0.047 | 0.090 | 0.180 | 0.195 | 0.176 | 0.251 | 0.177 | 0.239 | 0.140 | 0.164 | 0.205 | 0.196 | 0.193 |
| pEX.nTra10⋅SOD⋅His | 0.037 | 0.050 | 0.092 | 0.163 | 0.156 | 0.132 | 0.175 | 0.130 | 0.164 | 0.116 | 0.155 | 0.167 | 0.154 | 0.168 |
| pEX.nTAT⋅SOD⋅His | 0.033 | 0.046 | 0.090 | 0.170 | 0.177 | 0.161 | 0.225 | 0.153 | 0.225 | 0.045 | 0.170 | 0.162 | 0.193 | 0.200 |
| pEX.nLMWP⋅SOD⋅His | 0.031 | 0.042 | 0.86 | 0.174 | 0.192 | 0.178 | 0.237 | 0.170 | 0.242 | 0.137 | 0.180 | 0.185 | 0.200 | 0.200 |
| Dilution factor | 1 | 1 | 1 | 1 | 2 | 4 | 5 | 10 | 10 | 20 | 20 | 20 | 20 | 20 |
Results
Results indicate no significant difference in growth between different cultures. Slightly lower curve for the nTra10-expressing clone, which is somewhat interesting, since a similar progress was seen in the canceled experiment from 14/10. At least one more experiment will be run next week, using new BL21 clones; this will show whether this is just a coincidence or not.
Mimmi
clone non-CPP operon
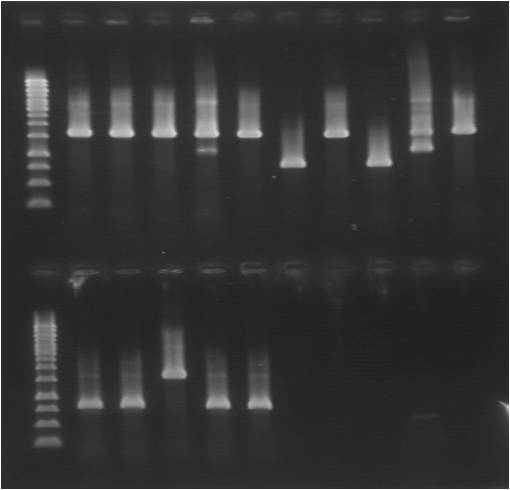
colony PCR
| mix | x16 | primers | samples | |||
|---|---|---|---|---|---|---|
| mastermix | 24.5 | pSB_VF2 | pSB1C3.SOD.RBS.yCCS 1-5 | |||
| DNA | 0.5 | pSB_VR | pSB1C3.SOD.his.RBS.yCCS 1-5 | |||
| tot | 25µl | pSB1C3.his.SOD.RBS.yCCS 1-5 | ||||
| positive control (pSB1C3.LMWP.SOD.his) | ||||||
Gel
| well | sample | well | sample |
|---|---|---|---|
| 1 | ladder | 12 | ladder |
| 2 | pC.S_yC 1 | 13 | pC.hS_yC 1 |
| 3 | pC.S_yC 2 | 14 | pC.hS_yC 2 |
| 4 | pC.S_yC 3 | 15 | pC.hS_yC 3 |
| 5 | pC.S_yC 4 | 16 | pC.hS_yC 4 |
| 6 | pC.S_yC 5 | 17 | pC.hS_yC 5 |
| 7 | pC.Sh_yC 1 | 18 | pC.L.hS, control |
| 8 | pC.Sh_yC 2 | ||
| 9 | pC.Sh_yC 3 | ||
| 10 | pC.Sh_yC 4 | ||
| 11 | pC.Sh_yC 5 |
ON culture
3ml LB Cm25 + toothpick from colony
SOD.RBS.yCCS 5 SOD.his.RBS.yCCS 2 his.SOD.RBA.yCCS 3
 |

|
 |

|
 |

|

|

|
 "
"