Team:Harvard/fences/design
From 2010.igem.org
| (13 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
<tr> | <tr> | ||
| - | <td | + | <td> |
| - | <div>Genetic Fence | + | <div>Genetic Fence Figure 2 <a href="http://openwetware.org/images/6/6c/Flowchart_Jamboree_slide.jpg" id="single_image" style="font-size:12px">click to enlarge</a></div><hr/> |
| - | <a href="http://openwetware.org/images/ | + | <a href="http://openwetware.org/images/6/6c/Flowchart_Jamboree_slide.jpg" id="single_image"> |
| - | <img src="http://openwetware.org/images/ | + | <img src="http://openwetware.org/images/6/6c/Flowchart_Jamboree_slide.jpg" width="300px" border=0> |
</a> | </a> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</td> | </td> | ||
</tr> | </tr> | ||
| Line 38: | Line 26: | ||
<p> | <p> | ||
| - | <b> | + | <b>fatal gene</b> |
<br> | <br> | ||
| - | A protein called Barnase prevents plants from growing outside the fence. Barnase, a non-specific RNAse, cleaves all pieces of RNA it encounters in the cell, thus derailing the cell’s metabolism and causing it to die. Barnase is expressed constitutively in all iGarden plants, so in the absence of Methoxyfenozide none will grow. | + | <p> |
| + | A protein called <a href="http://en.wikipedia.org/wiki/Barnase">Barnase</a> prevents plants from growing outside the fence. Barnase, a non-specific RNAse, cleaves all pieces of RNA it encounters in the cell, thus derailing the cell’s metabolism and causing it to die. Barnase is expressed constitutively in all iGarden plants, so in the absence of Methoxyfenozide none will grow. | ||
| + | </p> | ||
<br> | <br> | ||
<br> | <br> | ||
| - | <b> | + | <b>inhibitor</b> |
<br> | <br> | ||
| - | A protein called Barstar inhibits Barnase. When expressed in cells, Barstar prevents Barnase from destroying the iGarden plants. By putting Barstar under control of the switch-activated promoter (5xGal4 Upstream Activating Sequence plus 35s minimal promoter), barstar is expressed in the presence of | + | <p> |
| + | A protein called <a href="http://en.wikipedia.org/wiki/Barstar">Barstar</a> inhibits Barnase. When expressed in cells, Barstar prevents Barnase from destroying the iGarden plants. By putting Barstar under control of the switch-activated promoter (5xGal4 Upstream Activating Sequence plus 35s minimal promoter), barstar is expressed in the presence of Methoxyfenozide. | ||
| + | </p> | ||
| + | <p> | ||
| + | In addition to protein inhibition, a second switch-activated promoter regulates expression of a Lac Inhibitor Protein modified with a nuclear localization signal (LacIN). LacIN halts the transcription of the otherwise constitutive Actin2 promoter modified to include LacO sites (Act2LacO promoter). Barnase is placed under control of Act2LacO, so that it is expressed unless LacIN is present as well. Through LacIN and Barstar, Barnase is inhibited at both the transcriptional and the protein levels in the cell. | ||
<br> | <br> | ||
<br> | <br> | ||
| - | <b> | + | <b>sensor</b> |
<br> | <br> | ||
| - | The | + | <p> |
| + | The sensor component of the fence activates gene transcription in the presence of the fence compound Methoxyfenozide. Two receptor proteins, Ecdysone (EcR) and Retinoic Acid (RXR), form a dimer in the presense of methoxyfenozide. We created two fusion proteins, one combining Ecdysone with VP16 activating domain (which recruits the machinery to start transcription), and one combining Retinoic Acid with Gal4 DNA binding domain (which binds to a specific stretch of DNA where transcription will begin). When Methoxyfenozide dimerizes Ecdysone and Retinoic Acid, VP16 activating domain and Gal4 DNA binding domain act together to initiate transcription of Barstar and LacIN. | ||
| + | </p> | ||
<br> | <br> | ||
<br> | <br> | ||
| - | Thus, Methoxyfenozide | + | <p> |
| - | + | Thus, Methoxyfenozide activates the sensor, which results in Barstar and LacIN expression, which inhibits Barnase, allowing the plant to grow. | |
| + | </p> | ||
| Line 60: | Line 57: | ||
<div id="abstract"> | <div id="abstract"> | ||
<table style="padding:10px;color:#254117"> | <table style="padding:10px;color:#254117"> | ||
| - | + | <tr> | |
<td width="33%"> | <td width="33%"> | ||
| - | + | <!-- | |
<div>Genetic Fence Flowchart: Full <a href="http://openwetware.org/images/8/80/GeneticFenceFlowChart_Full.jpg" id="single_image" style="font-size:12px">click to enlarge</a></div><hr/> | <div>Genetic Fence Flowchart: Full <a href="http://openwetware.org/images/8/80/GeneticFenceFlowChart_Full.jpg" id="single_image" style="font-size:12px">click to enlarge</a></div><hr/> | ||
<a href="http://openwetware.org/images/8/80/GeneticFenceFlowChart_Full.jpg" id="single_image"> | <a href="http://openwetware.org/images/8/80/GeneticFenceFlowChart_Full.jpg" id="single_image"> | ||
<img src="http://openwetware.org/images/8/80/GeneticFenceFlowChart_Full.jpg" width="300px" border=0> | <img src="http://openwetware.org/images/8/80/GeneticFenceFlowChart_Full.jpg" width="300px" border=0> | ||
</a> | </a> | ||
| + | --> | ||
</td> | </td> | ||
| - | + | <div>Genetic Fence Design <a href="http://openwetware.org/images/f/fc/Updated_Jamboree_slides.jpg" id="single_image" style="font-size:12px">click to enlarge</a></div><hr/> | |
| + | <a href=" | ||
| + | http://openwetware.org/images/f/fc/Updated_Jamboree_slides.jpg" id="single_image"> | ||
| + | <img src=" | ||
| + | http://openwetware.org/images/f/fc/Updated_Jamboree_slides.jpg" width="300px" border=0> | ||
| + | </tr> | ||
<td width="33%"> | <td width="33%"> | ||
| - | < | + | <br /><br /> |
| - | + | ||
| - | + | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
</a> | </a> | ||
</td> | </td> | ||
| - | |||
</table> | </table> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 00:29, 27 October 2010

design
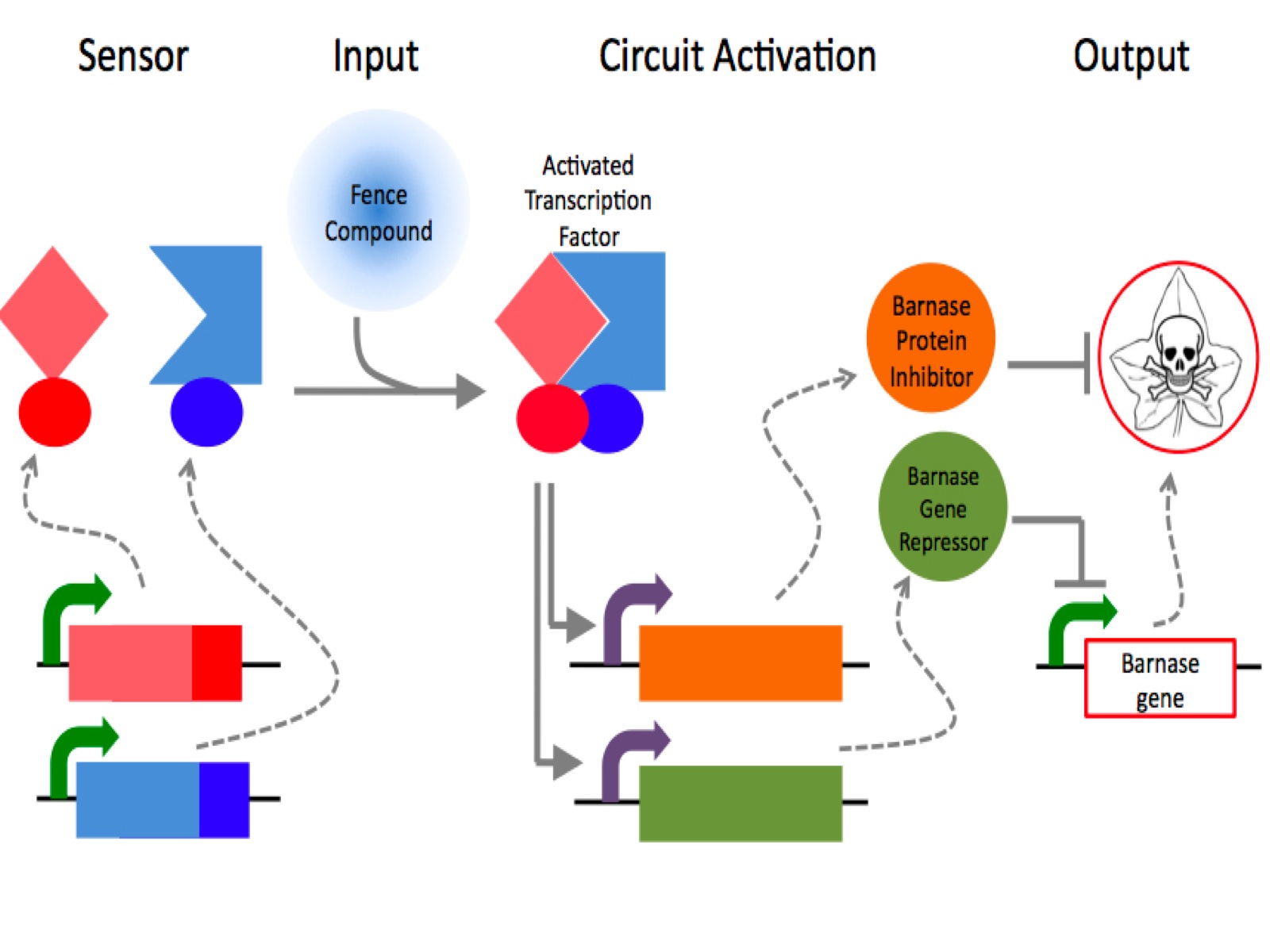
The genetic fence combines a chemically-activated gene switch with inhibition of a fatal gene pathway. In the presence of the fence compound, methoxyfenozide, the genetic switch activates transcription of an inhibitory mechanism, which halts an otherwise fatal gene pathway.
|
Genetic Fence Figure 2 click to enlarge 
|
fatal gene
A protein called Barnase prevents plants from growing outside the fence. Barnase, a non-specific RNAse, cleaves all pieces of RNA it encounters in the cell, thus derailing the cell’s metabolism and causing it to die. Barnase is expressed constitutively in all iGarden plants, so in the absence of Methoxyfenozide none will grow.
inhibitor
A protein called Barstar inhibits Barnase. When expressed in cells, Barstar prevents Barnase from destroying the iGarden plants. By putting Barstar under control of the switch-activated promoter (5xGal4 Upstream Activating Sequence plus 35s minimal promoter), barstar is expressed in the presence of Methoxyfenozide.
In addition to protein inhibition, a second switch-activated promoter regulates expression of a Lac Inhibitor Protein modified with a nuclear localization signal (LacIN). LacIN halts the transcription of the otherwise constitutive Actin2 promoter modified to include LacO sites (Act2LacO promoter). Barnase is placed under control of Act2LacO, so that it is expressed unless LacIN is present as well. Through LacIN and Barstar, Barnase is inhibited at both the transcriptional and the protein levels in the cell.
sensor
The sensor component of the fence activates gene transcription in the presence of the fence compound Methoxyfenozide. Two receptor proteins, Ecdysone (EcR) and Retinoic Acid (RXR), form a dimer in the presense of methoxyfenozide. We created two fusion proteins, one combining Ecdysone with VP16 activating domain (which recruits the machinery to start transcription), and one combining Retinoic Acid with Gal4 DNA binding domain (which binds to a specific stretch of DNA where transcription will begin). When Methoxyfenozide dimerizes Ecdysone and Retinoic Acid, VP16 activating domain and Gal4 DNA binding domain act together to initiate transcription of Barstar and LacIN.
Thus, Methoxyfenozide activates the sensor, which results in Barstar and LacIN expression, which inhibits Barnase, allowing the plant to grow.
|
|
 "
"