Team:Berkeley/Project/Overview
From 2010.igem.org
(New page: Based on our goals, we had to devise a way of delivering protein and DNA payloads to the cytoplasm of a choanoflagellate. During normal digestion the choanoflagellate will envelope a bacte...) |
|||
| (42 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:Overview header.png | 956px]] | |
| - | [[Image:BacteriaWithPayload.jpg]] | + | __NOTOC__ |
| - | [[Image: | + | <!-- Global templates. Should be included on every page --> |
| - | [[Image: | + | {{Berkeley_Main_Script}} |
| - | [[Image: | + | {{Berkeley_CSS}} |
| - | [[Image: | + | {{Berkeley_Menu}} |
| + | <!-- End global templates. Insert content below --> | ||
| + | |||
| + | This year our goal was to engineer <i>Escheria Coli</i> bacteria to serve as a vector for delivering protein and DNA payloads to the cytoplasms of phagocytic eukaryotes. As a proof of concept, we attempted to deliver a GFP payload to the cytoplasm of choanoflagellates. (See [https://2010.igem.org/Team:Berkeley/Project/Motivation Motivation] for more information on choice of choanoflagellates). We are excited to report that we have been able to achieve successful delivery of GFP to the cytoplasms of choanoflagellates with a high rate of delivery! Visit our [https://2010.igem.org/Team:Berkeley/Results Results] page for more information. | ||
| + | <br> | ||
| + | <br> | ||
| + | Our delivery scheme is not specific to either GFP or choanoflagellates, and is theoretically capable of delivering more complex [https://2010.igem.org/Team:Berkeley/Project/Payload payloads] to a variety of eukaryotic [https://2010.igem.org/Team:Berkeley/Project/Other_organisms organisms]. This work is the first step towards genetic manipulation of eukaryotes using bacteria equipped with BioBricks devices. | ||
| + | <br><br> | ||
| + | The design and execution of our project brought up issues of biosafety, which are discussed on our [https://2010.igem.org/Team:Berkeley/Project/Human_Practices Human Practices] page. In addition to the wet lab work outlined above, our team made significant improvements to the [https://2010.igem.org/Team:Berkeley/Clotho Clotho] software originally presented by the [https://2008.igem.org/Team:UC_Berkeley_Tools 2008 Berkeley iGEM Software team]. | ||
| + | <br> | ||
| + | <br> | ||
| + | An overview of our delivery mechanism is presented below: | ||
| + | ==Delivery Scheme== | ||
| + | {| cellpadding="10" cellspacing="10" | ||
| + | |<b>Payload</b><br> | ||
| + | We start with a bacteria that expresses a [https://2010.igem.org/Team:Berkeley/Project/Payload Payload], either proteins, nucleic acids, or a combination both and a payload delivery device. We define a payload delivery device as the combination of a [https://2010.igem.org/Team:Berkeley/Project/Self_Lysis Self-Lysis] part and a [https://2010.igem.org/Team:Berkeley/Project/Vesicle_Buster Vesicle-Buster] part. | ||
| + | ||<center> [[Image:BacteriaWithPayload.jpg|200px]] </center> | ||
| + | |- | ||
| + | |<b>Barriers to Delivery</b><br> | ||
| + | Choanoflagellates (and many other lower metazoans) naturally eat bacteria, so during normal digestion the choanoflagellate envelopes bacteria in a phagocytotic vesicle. When the bacteria is in the phagocytotic vesicle, there are two barriers between our payload and the cytoplasm of the choanoflagellate, the bacteria's own membranes, and the vesicle membrane itself. | ||
| + | ||[[Image:BacteriaInVesicleInChoano.jpg|500px]] | ||
| + | |- | ||
| + | |<b>Perils of Digestion</b><br> | ||
| + | This vesicle is then transported to choanoflagellate's food vacuole, where it merges with the vacuole. At this point the contents of the vesicle are exposed to the inside of the food vacuole and everything is destroyed, including the bacteria and all its contents. | ||
| + | || [[Image:ChoanoDigestingBacteria.jpg|500px]] | ||
| + | |- | ||
| + | |<b>Self-Lysis</b><br> | ||
| + | In order to achieve successful delivery, our bacteria must be able to deliver the payload in the short time between ingestion and digestion. Once our bacteria is engulfed by the choanoflagellate, and an inducer is added, it lyses itself using the [https://2010.igem.org/Team:Berkeley/Project/Self_Lysis Self-Lysis] device derived from the 2008 UC Berkeley iGEM team. | ||
| + | ||[[Image:LysingBacteriaInChoano.jpg|500px]] | ||
| + | |- | ||
| + | |<b>Vesicle-Buster</b><br> | ||
| + | At this point, the proteins being expressed by the bacteria will be released into the vesicle. Included in these proteins is the [https://2010.igem.org/Team:Berkeley/Project/Vesicle_Buster Vesicle-Buster] device, which punctures the food vesicle membrane and releases the payload into the cytoplasm of the choanoflagellate. | ||
| + | ||[[Image:BustingVesicleInChoano.jpg|500px]] | ||
| + | |} | ||
Latest revision as of 06:02, 27 October 2010
- Home
- Project
- Overview
- Motivation
- Other Organisms
- Parts
- Self-Lysis
- Vesicle-Buster
- Payload
- [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=Berkeley Parts Submitted]
- Results
- Judging
- Clotho
- Human Practices
- Team Resources
- Who We Are
- Notebooks:
- [http://www.openwetware.org/wiki/Berk2010-Daniela Daniela's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Christoph Christoph's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Amy Amy's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Tahoura Tahoura's Notebook]
- [http://www.openwetware.org/wiki/Berk2010-Conor Conor's Notebook]
This year our goal was to engineer Escheria Coli bacteria to serve as a vector for delivering protein and DNA payloads to the cytoplasms of phagocytic eukaryotes. As a proof of concept, we attempted to deliver a GFP payload to the cytoplasm of choanoflagellates. (See Motivation for more information on choice of choanoflagellates). We are excited to report that we have been able to achieve successful delivery of GFP to the cytoplasms of choanoflagellates with a high rate of delivery! Visit our Results page for more information.
Our delivery scheme is not specific to either GFP or choanoflagellates, and is theoretically capable of delivering more complex payloads to a variety of eukaryotic organisms. This work is the first step towards genetic manipulation of eukaryotes using bacteria equipped with BioBricks devices.
The design and execution of our project brought up issues of biosafety, which are discussed on our Human Practices page. In addition to the wet lab work outlined above, our team made significant improvements to the Clotho software originally presented by the 2008 Berkeley iGEM Software team.
An overview of our delivery mechanism is presented below:
Delivery Scheme
| Payload We start with a bacteria that expresses a Payload, either proteins, nucleic acids, or a combination both and a payload delivery device. We define a payload delivery device as the combination of a Self-Lysis part and a Vesicle-Buster part. | 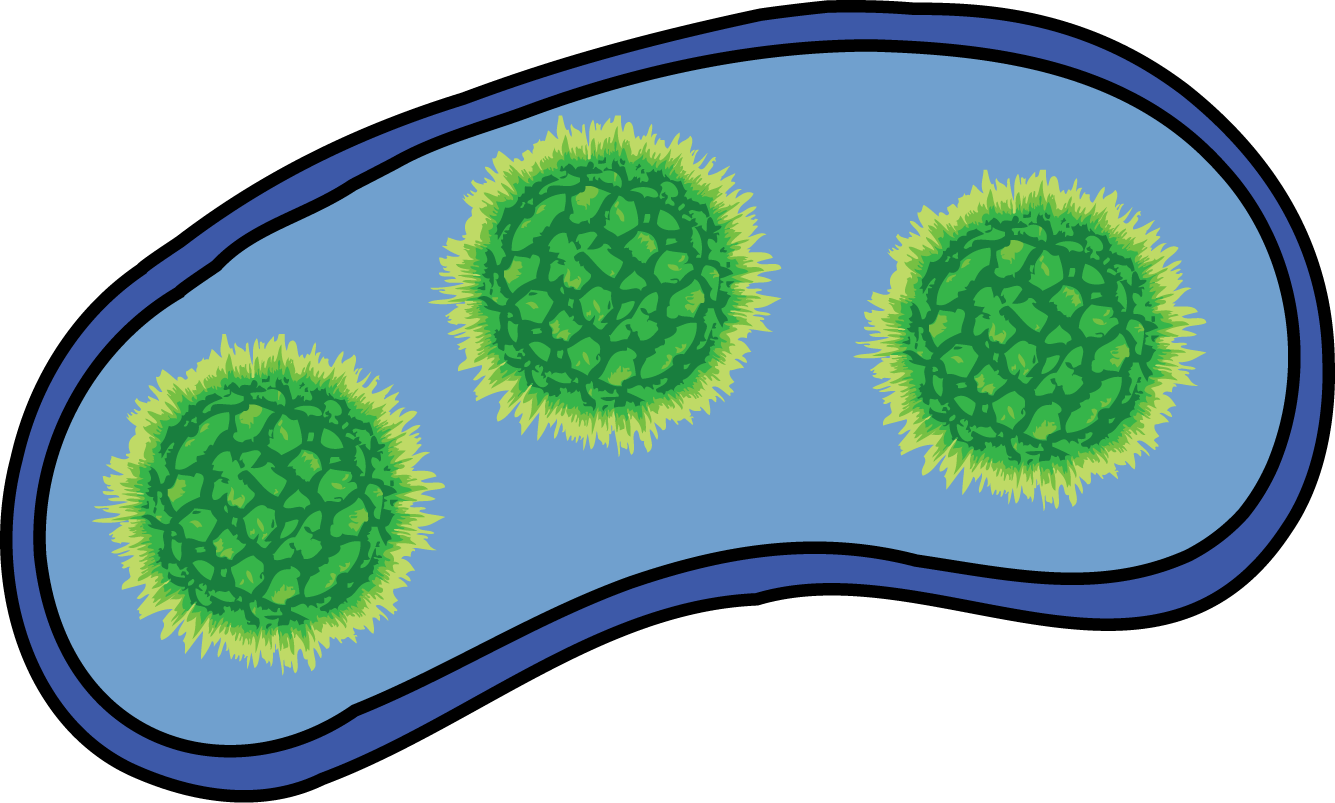 |
| Barriers to Delivery Choanoflagellates (and many other lower metazoans) naturally eat bacteria, so during normal digestion the choanoflagellate envelopes bacteria in a phagocytotic vesicle. When the bacteria is in the phagocytotic vesicle, there are two barriers between our payload and the cytoplasm of the choanoflagellate, the bacteria's own membranes, and the vesicle membrane itself. | 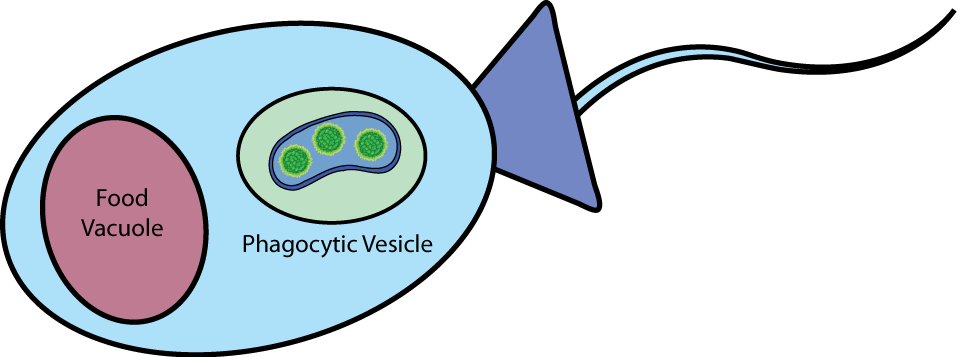
|
| Perils of Digestion This vesicle is then transported to choanoflagellate's food vacuole, where it merges with the vacuole. At this point the contents of the vesicle are exposed to the inside of the food vacuole and everything is destroyed, including the bacteria and all its contents. | 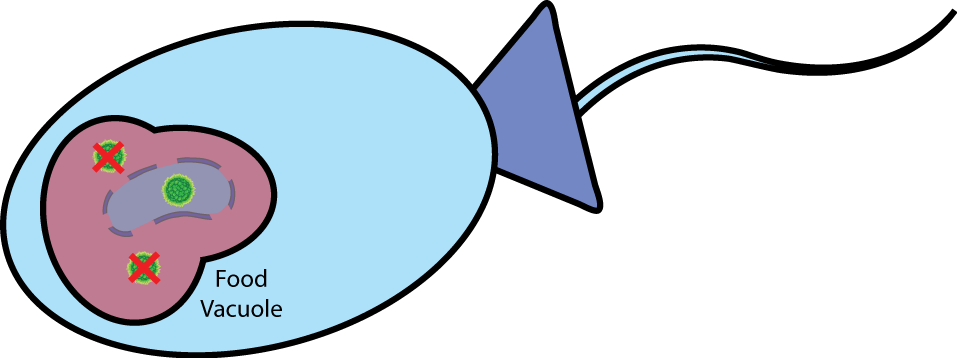
|
| Self-Lysis In order to achieve successful delivery, our bacteria must be able to deliver the payload in the short time between ingestion and digestion. Once our bacteria is engulfed by the choanoflagellate, and an inducer is added, it lyses itself using the Self-Lysis device derived from the 2008 UC Berkeley iGEM team. | 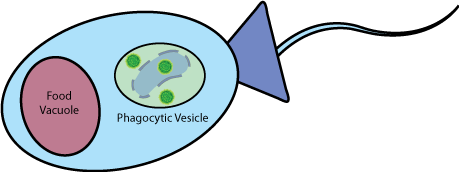
|
| Vesicle-Buster At this point, the proteins being expressed by the bacteria will be released into the vesicle. Included in these proteins is the Vesicle-Buster device, which punctures the food vesicle membrane and releases the payload into the cytoplasm of the choanoflagellate. | 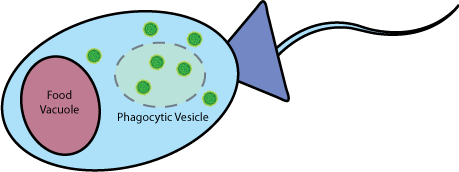
|
 "
"
