Team:LMU-Munich/Jump-or-die/Functional Principle
From 2010.igem.org
(Difference between revisions)
(→Jump-or-die-System) |
|||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:LMU-Munich/Templates/Page Header}} | {{:Team:LMU-Munich/Templates/Page Header}} | ||
| + | |||
| + | [[image:Jumplogo.png|250px|right|ApoControl logo]] | ||
| + | |||
== Jump-or-die-System == | == Jump-or-die-System == | ||
| + | |||
The Jump-or-Die-system facilitates the integration of a gene of interest into cellular genome and selects the cells successfully integrated with the gene of interest by apoptosis. Therefore three constructs are needed. | The Jump-or-Die-system facilitates the integration of a gene of interest into cellular genome and selects the cells successfully integrated with the gene of interest by apoptosis. Therefore three constructs are needed. | ||
| Line 7: | Line 11: | ||
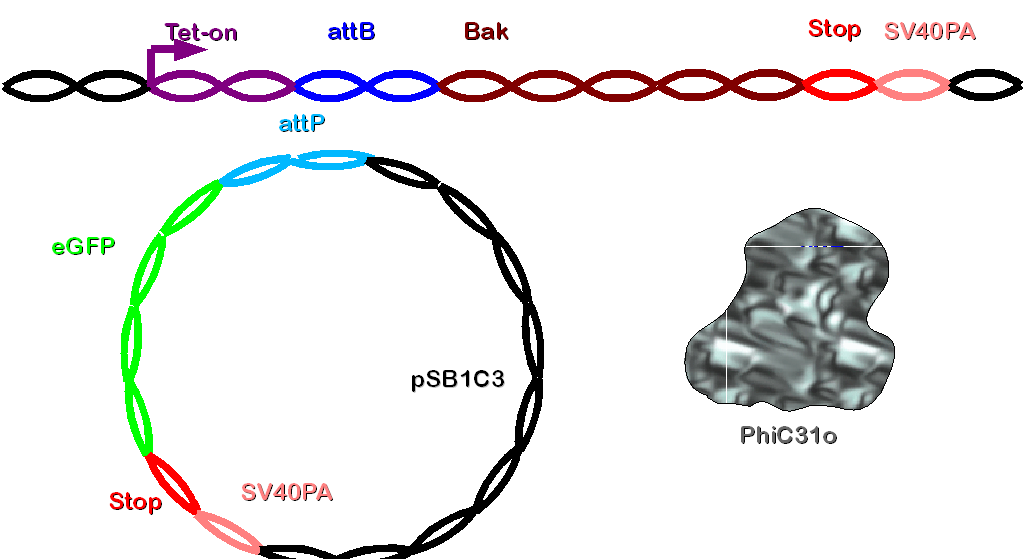
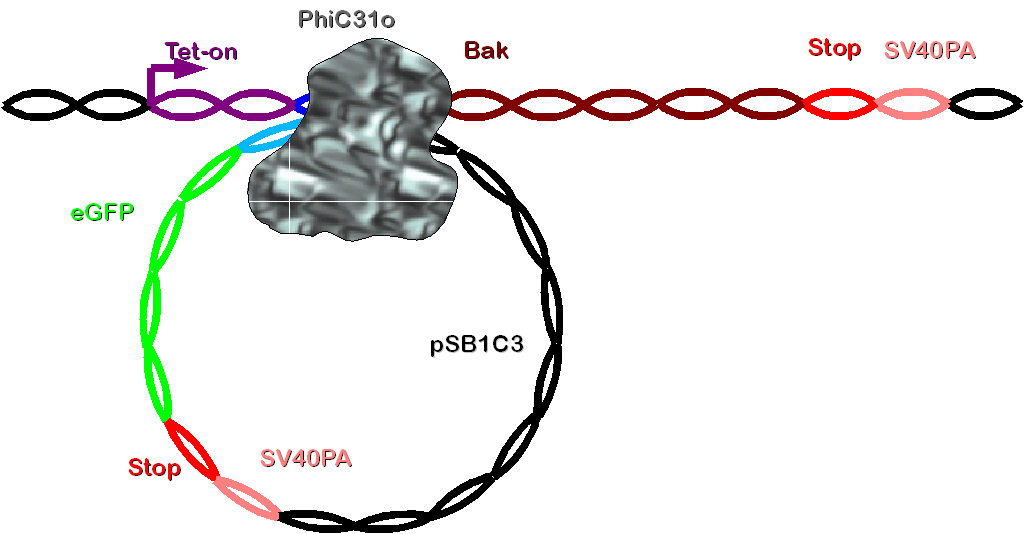
Construct 1 contains a tet-on promoter, bacterial attachment site (attB), human bak (bak) and SV40 polyadenylation site (SV40PA). It has to be integrated to the genome of the cellline. The selection of the cells stably transfected with this construct is realized by co-transfection with hygromycine resistance. | Construct 1 contains a tet-on promoter, bacterial attachment site (attB), human bak (bak) and SV40 polyadenylation site (SV40PA). It has to be integrated to the genome of the cellline. The selection of the cells stably transfected with this construct is realized by co-transfection with hygromycine resistance. | ||
| - | [[Image: | + | [[Image: Dnajump1.PNG | 600px | Jump construct 1]] |
=== Construct 2 === | === Construct 2 === | ||
| Line 13: | Line 17: | ||
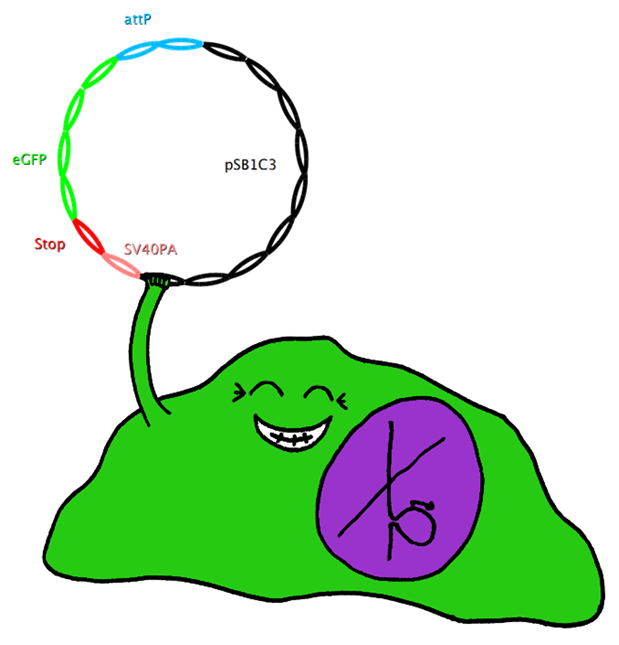
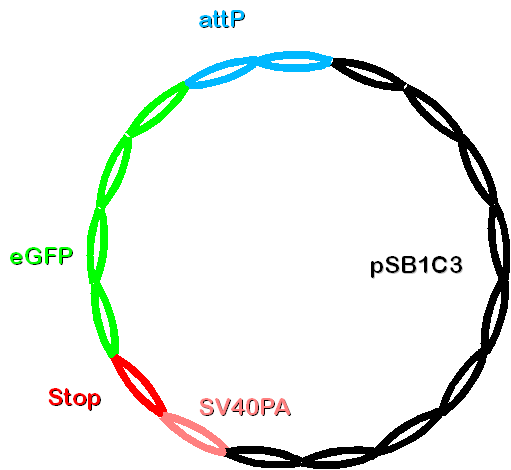
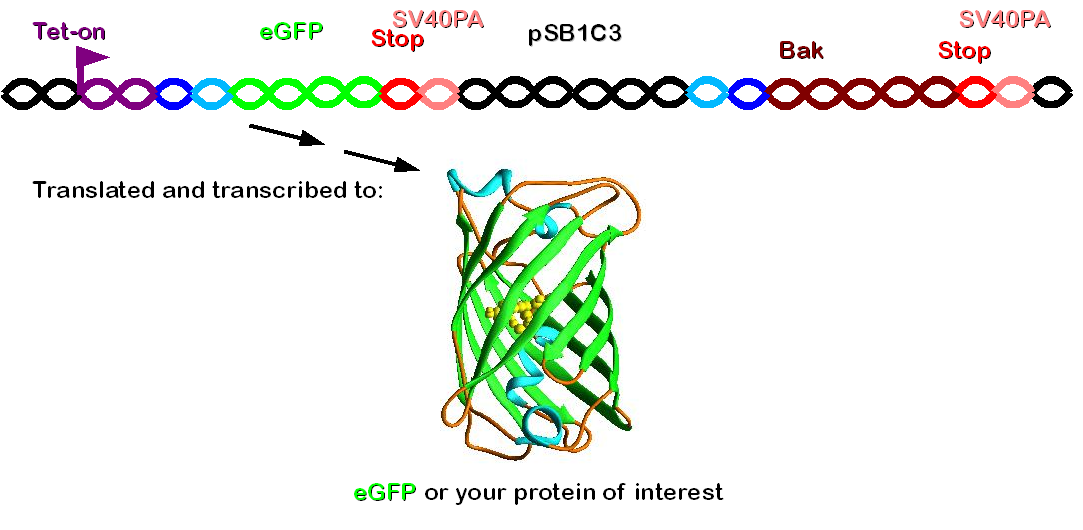
Construct 2 consists of the following parts: phage attachment site (attP), eGFP (our gene of interest), doubled stop codon and SV40PA. | Construct 2 consists of the following parts: phage attachment site (attP), eGFP (our gene of interest), doubled stop codon and SV40PA. | ||
| - | [[Image: | + | [[Image: Dnajump2.PNG | 200px | Jump construct 2]] |
=== Construct 3 === | === Construct 3 === | ||
| Line 19: | Line 23: | ||
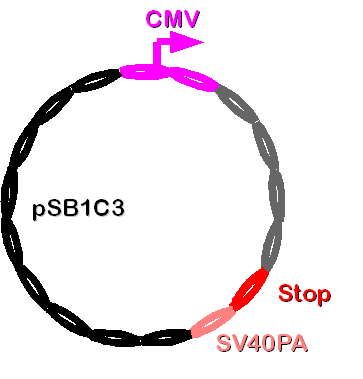
This Biobrick contains CMV-promoter, PhiC31o (mouse-codon optimized integrase) and SV40PA. It will be transfected to the cellline and read-off so that PhiC31o can catalyze the integration of Construct 2 into the attB site in genome. | This Biobrick contains CMV-promoter, PhiC31o (mouse-codon optimized integrase) and SV40PA. It will be transfected to the cellline and read-off so that PhiC31o can catalyze the integration of Construct 2 into the attB site in genome. | ||
| - | [[Image: | + | [[Image: Dnajump3.PNG | 200px | Jump construct 3]] |
== Transfection possibilities == | == Transfection possibilities == | ||
| - | A cellline which has construct 1 integrated into the genome will be co-transfected with construct 2 and 3. Construct 3 will be read-off instantly, in contrast to construct 1 which has to be induced by | + | A cellline which has construct 1 integrated into the genome will be co-transfected with construct 2 and 3. Construct 3 will be read-off instantly, in contrast to construct 1 which has to be induced by doxycycline and because of the lack of a promoter, construct 2 will at the beginning not be translated. |
The outgoing of this co-transfection has the following possibilities: | The outgoing of this co-transfection has the following possibilities: | ||
| Line 50: | Line 54: | ||
=== d) both constructs 2 and 3 enter the cell === | === d) both constructs 2 and 3 enter the cell === | ||
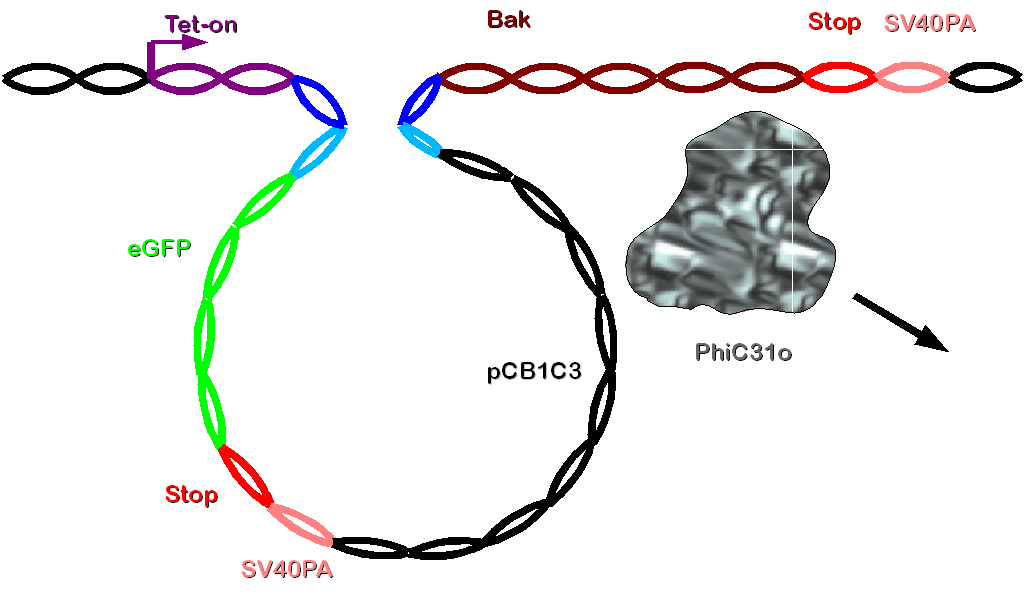
| - | PhiC31o will be read off because of CMV promoter. Now the integrase will combine attB and attP site and integrate construct 2 into the genome. Now the gene of interest is integrated between the tet-on promoter and bak in the cellular genome. This complex will be translated after | + | PhiC31o will be read off because of CMV promoter. Now the integrase will combine attB and attP site and integrate construct 2 into the genome by a specific rate. All cells where PhiC31o doesn't integrate contruct 2 will also die due to Bak after doxycycline induction Now the gene of interest is integrated between the tet-on promoter and bak in the cellular genome. This complex will be translated after doxycycline induction. The doubled stop codon and polyadenylation site directly after eGFP (the gene of interest) will efficiently prevent the translation of bak. Thus such cells will express the gene of interest and survive. |
| + | |||
| + | [[Image: Dnajump4.PNG | 600px | Jump recombination]] | ||
| + | |||
| + | [[Image: Dnajump8.PNG |center| 200px | Jump recombination]] | ||
| + | |||
| + | [[Image: Dnajump5.PNG | 600px | Jump recombination]] | ||
| + | |||
| + | [[Image: Dnajump8.PNG |center| 200px | Jump recombination]] | ||
| + | |||
| + | [[Image: Dnajump6.PNG | 600px | Jump recombination]] | ||
| - | [[Image: | + | [[Image: Dnajump8.PNG |center| 200px | Jump recombination]] |
| + | [[Image: Dnajump7.PNG | 600px | Jump recombination]] | ||
| + | [[image:Jumplogo.png|250px|right|ApoControl logo]] | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 14:45, 25 October 2010


![]()
![]()







![]()
 "
"