Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
Laura Nadine (Talk | contribs) (→Introduction) |
(→miTuner Kit components) |
||
| (134 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| - | + | ||
<html> | <html> | ||
| + | <a name="top"></a> | ||
<style type="text/css"> | <style type="text/css"> | ||
.tocnumber {display:none;} | .tocnumber {display:none;} | ||
| - | |||
#toc ul ul, .toc ul ul { | #toc ul ul, .toc ul ul { | ||
margin:0 0 0 0em; | margin:0 0 0 0em; | ||
| - | |||
} | } | ||
| - | |||
#toctitle {display:none;} | #toctitle {display:none;} | ||
| - | |||
</style> | </style> | ||
</html> | </html> | ||
| + | {{:Team:Heidelberg/Single_Pagetop|project_miRNA_Kit}} | ||
| + | {{:Team:Heidelberg/Side_Top}} | ||
| + | __NOTOC__ | ||
| - | + | <br/> | |
| + | <center> | ||
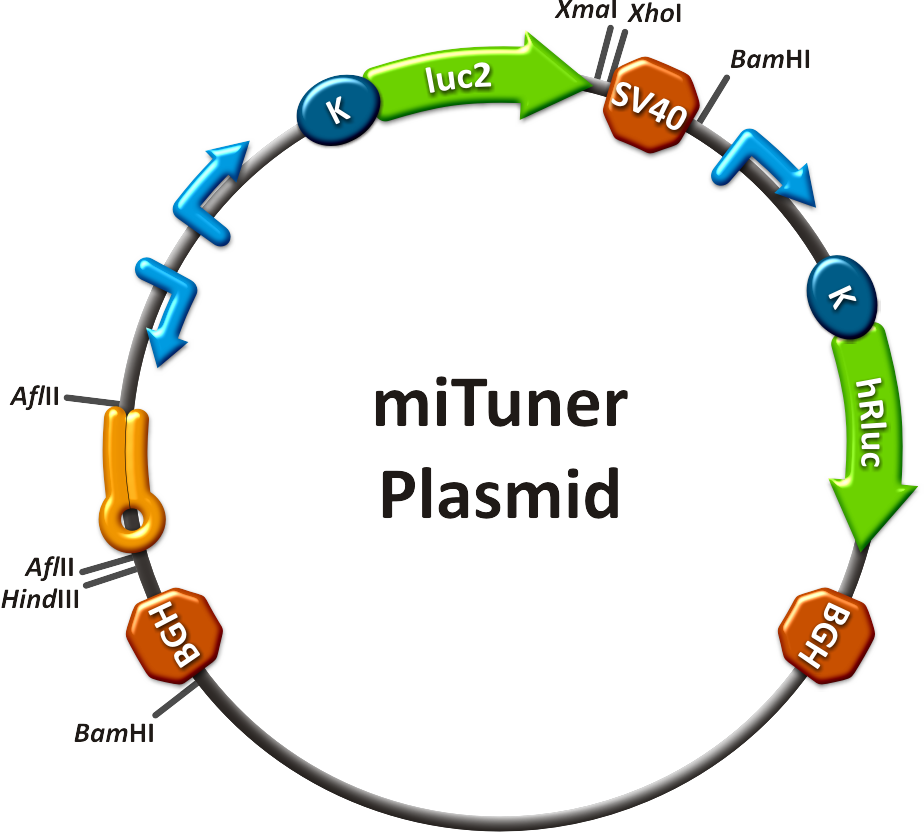
| + | [[Image:MiTuner p.png|250px| miTuner plasmid]] | ||
| + | </center> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | === Working Modes === | ||
| + | The synthetic miR Kit can be applied in three different ways: | ||
| + | :I) '''Tuning''': adjusting the expression <br/>of the GOI by expressing a synthetic microRNA in the target cell/tissue | ||
| + | <br/> | ||
| + | :II) '''Off-Targeting''': switching OFF the expression <br/>of the GOI in case a certain endogenous microRNA is present in the target cell/tissue | ||
| + | <br/> | ||
| + | :III) '''On-Targeting''': switching ON the expression <br/>of the GOI in case a certain endogenous microRNA is present in the target cell/tissue | ||
| + | <br/> | ||
| + | |||
| + | |||
| + | === Simple Tuning Procedure === | ||
| + | * choose an [[Team:Heidelberg/Project/Introduction | interesting microRNA]] | ||
| + | * [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner create] referring binding sites | ||
| + | * order your binding site oligos | ||
| + | * [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Cloning clone] them into your [https://2010.igem.org/Team:Heidelberg/Parts#final_constructs miTuner construct] | ||
| + | * [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Transfection transfect] your cells | ||
| + | * measure the [[Team:Heidelberg/Project/miRNA Kit#Results | tuned]] expression! | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | === Advancement === | ||
| + | * digestion of miR Kit construct with BamHI | ||
| + | * cloning into viral backbone (e. g. [https://2010.igem.org/Team:Heidelberg/Notebook/Material pBS_U6]) | ||
| + | * [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Virus_Production virus production] | ||
| + | * infection of cells | ||
| + | * achievement of specific target cell [https://2010.igem.org/Team:Heidelberg/Project/Capsid_Shuffling tropism] | ||
| + | → further improvement of gene expression tuning | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | |||
| + | === Tuning Raw Data === | ||
| + | For our <i>in vitro</i> tuning, you can have a look even at our unprocessed data with specific [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#nomenclature nomenclature]: | ||
| + | * [[Media:Plate1 process H1.xls]], <br/> | ||
| + | * [[Media:Plate2 process H1.xls]], <br/> | ||
| + | * [[Media:Plate3 process H1.xls]], <br/> | ||
| + | * [[Media:Haat 20101022 M1-M4 ctrl H1.xls]], <br/> | ||
| + | * [[Media:Haat 20101026 M5-M8 ctrl H1.xls]], <br/> | ||
| + | * [[Media:Haat 20101026 M9M22 ctrl H1.xls]], <br/> | ||
| + | * [[Media:HAAT H1 final.xls]]. <br/> | ||
| + | *[[Media:Plate1 process U6 haat.xls]],<br/> | ||
| + | *[[Media:Plate2 process U6 haat.xls]],<br/> | ||
| + | *[[Media:Plate3 process U6 haat.xls]],<br/> | ||
| + | *[[Media:Haat 20101026 plate2 U6.xls]],<br/> | ||
| + | *[[Media:Haat 20101026 plate1 U6.xls]],<br/> | ||
| + | *[[Media:Haat 20101026 plate3 U6.xls]],<br/> | ||
| + | *[[Media:HAAT U6 final.xls]].<br/> | ||
| + | {{:Team:Heidelberg/Side_Bottom}} | ||
<div class="t1">Synthetic miRNA Kit</div> | <div class="t1">Synthetic miRNA Kit</div> | ||
| + | |||
| + | <center><h4>miTuner - a kit for microRNA based gene expression tuning in mammalian cells</h4></center> | ||
| + | <br/> | ||
| + | <center><i>With the synthetic miRNA kit, we provide a comprehensive mean | ||
| + | to plan, conduct and evaluate experiments dealing with [https://2010.igem.org/Team:Heidelberg/Parts miBricks] | ||
| + | (i. e. microRNA related Biobricks) as key regulators in mammalian cells.</i></center> | ||
==Abstract== | ==Abstract== | ||
| + | |||
| + | Regulation of any gene of interest has never been as easy as with our '''miRNA-based expression tuning kit miTuner'''. Rational design of synthetic miRNA binding sites according to our recommendations enables fine-tuning of gene expression in a range between 5% and 100%. Additionally, we offer Off- and On-targeting switches which effect GOI expression in only one or all but one tissue depending on endogenously occurring miRNAs. | ||
| + | [[Image:Mitunerabstract.png|thumb|370px|left|'''Figure 1''': Three modes of gene regulation accomplished by miTuner. '''a)'''fine-tuning of gene expression. '''b) and c)''' Off- and On-targeting for tissue specific expression of a therapeutic gene]] | ||
| + | The '''tuning application''' is based on a dual promoter construct that expresses a GOI controlled by a synthetic miRNA which is expressed from either the same or a second construct ('''figure 1a'''). Differing miRNA-binding site interaction efficiencies caused by binding sites of different sequence properties are used to distinctly adjust expression strength of the GOI. | ||
| + | |||
| + | For '''Off-targeting''', the GOI is under control of miRNAs that are found in tissue where gene expression is thereupon silenced while the GOI can still be expressed in other tissues as visualized in '''figure 1b'''. | ||
| + | |||
| + | '''On-targeting''' is based on the expression of the GOI from a promoter containing a Tet Operator (Tet02) that negatively regulates gene expression in the presence of a Tet Repressor ('''figure 1c'''). If the Tet Repressor tagged with binding sites for an endogenous miRNA, that is specifically expressed in the target cells/tissue. In consequence, the TetR is knocked down, releasing the promoter and enabling specific GOI expression. | ||
| + | |||
==Introduction== | ==Introduction== | ||
| - | MicroRNAs (miRNAs) are endogenous | + | MicroRNAs (miRNAs) are short endogenous, non-coding RNAs that mediate gene expression in a diversity of organisms {{HDref|Bartel, 2004}}. Although the understanding of their biological functions is progressing remarkably, the exact mechanisms of regulation are still not unambiguously defined. However, it is commonly believed that miRNAs '''trigger target mRNA regulation''' by binding to 3’ untranslated region (UTR) of its target {{HDref|Chekulaeva and Filipowicz, 2009}}. <!--The discovery of the first miRNA (lin-4) revealed sequence complementarity to multiple conserved sites in the 3’UTR of the lin-14 mRNA {{HDref|Lee et al., 1993; Wightman et al., 1993}}. --> Exact principles of expression knockdown mediated by miRNA are still in debate {{HDref|Eulalio et al., 2008}}.<br/>However, sequence depending '''binding site properties''' have an essential impact on miRNA-mRNA interaction. <!--[figure, short explanations on seed regions, flanking regions, spacers, mismatches and resulting bulges]. Some functionally important sections of miRNAs have been described in literature, such as the seed region {{HDref|Grimson et al., 2007; Bartel, 2009}}. It is defined as a miRNA region of seven nucleotides length that shows perfect pairing the mRNA target sequence. --><!--The seed usually consists of the nucleotides on position 2-8 of a miRNA binding sites in the 5'UTR of the mRNA. Based on this simple principle, we randomized our miRNA binding sites between nucleotide 9 - 12 or 9 - 22 in the so called flanking region. Alternatively, we tried rational exchanges of nucleotides to see how they effect binding of the miRNA to its target mRNA. --> Depending on pairing specificity translational repression is mediated through the imperfect miRNA-mRNA hybrids. The potential for stringent regulation of transgene expression makes the miRNA world a promising area of gene therapy {{HDref|Brown et al.,2009}}. There is a need for tight control of gene expression, since cellular processes are sensitive to expression profiles. Non-mediated gene expression can lead to fatal dysfunction of molecular networks. It is widely known, that miRNAs can adjust such fluctuations {{HDref|Brenecke et al., 2005}}. A combination of random and rational '''design''' of binding sites could become a '''powerful tool''' to achieve a narrow range of resulting gene expression knockdown. To ease <i>in silico</i> construction of miRNA binding sites with appropriate characteristics for its target, we wrote a program - the [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner miBS designer]. Using all of our [https://2010.igem.org/Team:Heidelberg/Modeling theoretical models] gives the user the opportunity to calculate knockdown percentages caused by the designed miRNA in the target cell.<!--The experimental applicability is still limited by redundant target sites and various miRNA expression patterns within the cells. This hampers distinct expression levels of the gene of interest (GOI) fused to the miRNA binding site.--> |
| + | Our '''synthetic miRNA Kit''' guarantees at least for individually modifiable but still ready-to-use constructs to interfere genetic circuits with synthetic or endogenous miRNAs. We preciously show, that gene expression can thereby by adjusted - tuned - to an arbitrary level. The '''miTuner''' (see sidebar) allows on the simultaneous expression of a synthetic miRNA and a gene of interest that is fused with a designed binding site for this specific miRNA. Our modular kit comes with different parts that can be combined by choice, e. g. different mammalian promoters and characterized binding sites of specific properties. By choosing a certain binding site to tag the GOI, one can tune the expression of this gene. Depending on the GOI, different means for read out of gene expression come into play. At first, we applied [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay dual-luciferase assay], since we used Luciferase as a reporter for a proof-of-principle approach. Later on, semi-quantitative immunoblots were prepared for testing of therapeutic genes. However, all the received information fed our models, thereby creating an '''integrative feedback loop between experiments and in silico simulation'''. | ||
| - | + | === miTuner Kit components === | |
| + | The miTuner Kit consists of three basic components: <br /> | ||
| + | :a) A kit of standardized synthetic microRNAs, corresponding binding sites, promoters and luciferase expression constructs as well as measurement constructs assembled in the BB-2 standard. As the miTuner kit was enabled <br /> | ||
| + | :b) Protocols for engineering synthetic microRNAs, synthetic single microRNA binding sites as well as microRNA binding site patterns | ||
| + | Please find basic information about the kit components and engineering of the kit [[Team:Heidelberg/Project/miRNA Kit_engineer|here]] | ||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
==Results== | ==Results== | ||
| + | |||
| + | All gene regulatory constructs for tuning, Off- or On-targeting can easily be assembled using '''BBB standard cloning''' from our miRNA Kit [https://2010.igem.org/Team:Heidelberg/Parts parts]. After successful cloning, the constructs can be transfected onto a cell line of choice or transferred into a virus backbone for [https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection ''in vivo''] experiments. For our '''proof of principle''', we used firefly luciferase normalized to ''Renilla'' luciferase on miTuner to characterize knockdown efficiencies of different binding sites and show Off- and On-targeting by mouse infection carried by an AAV virus. | ||
| + | |||
| + | ===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== | ||
| + | |||
| + | The data shows a precisely tuned expression from almost 0% to 100% (Fig. 2, Fig. 3). Lowest expression refers to complete knockdown through fusion of perfect binding sites (always green bar on the left hand side of the figures) to the reporter gene. Expression from a construct without binding sites is set as 100% (always orange column on the right hand side of the figures). In presence of the specific shRNA miR, gene expression was mediated to various levels through interactions with the different imperfect binding sites. Whereas, when an unspecific shRNA miR was expressed, gene expression remained unaffected (see raw data below). This reference shows that the binding sites were correctly designed, since they seem to interact specifically with a referring shRNA miR. | ||
| + | |||
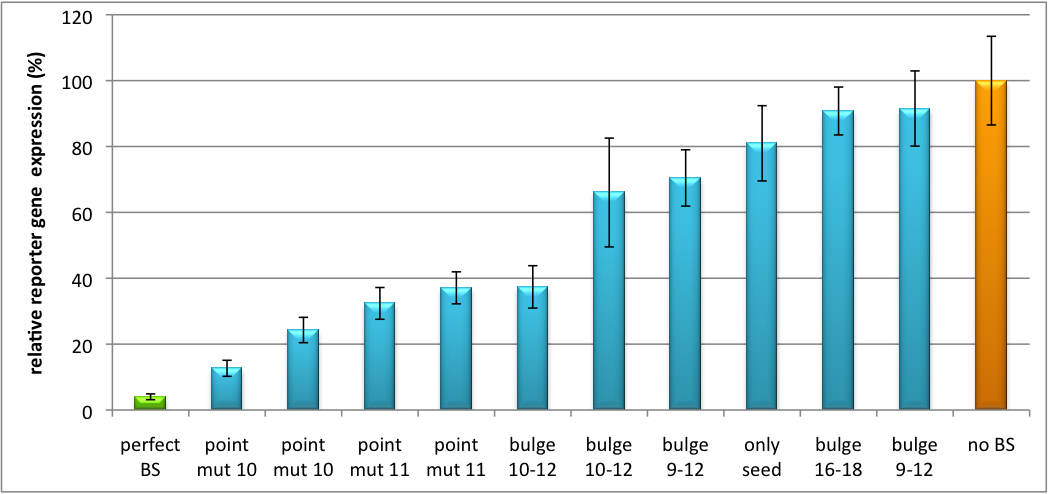
| + | [[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure 2: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_SV40 Luc2 double transfected with a reference renilla construct.''' The shRNA_hAAT construct was expressed from a pBS_H1 construct.]] | ||
| + | |||
| + | Figure 2 shows the results of Dual-Luciferase measurements of the miTuner plasmid with binding sites against shhAAT behind firefly luciferase. The highest knockdown can be achieved by using a perfect binding site. Single mutations outside the seed region at position 11, 12 or 10-12 lead to knockdown between 10% and 60% compared to unregulated expression. Bulges close to the seed region or changes in the seed region itself lead to very low downregulation. Having only the seed region as a target for the miRNA also leads to a less efficient knockdown compared with binding sites containing flanking regions. | ||
| + | |||
| + | [[Image:Haat_U6HD2010.jpg|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_SV40_Luc2 construct cotransfected with a reference renilla construct.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT. The shhAAT was expressed from a pBS_U6 plasmid]] | ||
| + | |||
| + | Figure 3 shows the same assay using binding sites against shhAAT. This time, the shhAAT is driven by a U6 promoter, which is stronger than the H1 promoter used for driving the shRNA in the previous figure. The results are overall similar, with changes in or directly adjacent to the seed region having the highest impact on knockdown efficiency. | ||
| + | |||
| + | [[Image:PsiCheck.png|thumb|center|600px|'''Figure 4: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | ||
| + | |||
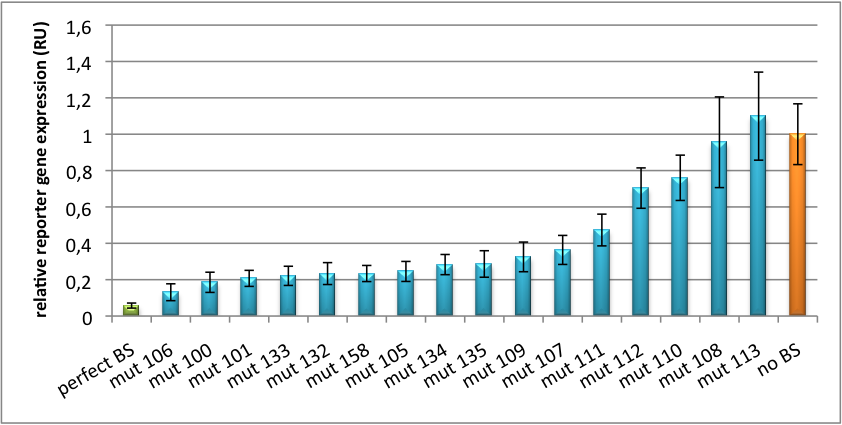
| + | We further analyzed binding sites derived from miR122 in the dual luciferase vector PsiCheck2 as can be seen in figure 4. Here we tested sixteen mutated binding sites in order to observe minute fine-tuning between one binding site and the next. Mutated Binding sites 123, 133, 134, 135 and 158 contain 4bp-bulges (non-paired regions) that don not seem to diminish knockdown efficiency much. 107 contains one binding site, while 134 and 135 contain two binding sites for the same miRNA and show a stronger knockdown than 107. | ||
| + | |||
| + | ===Off-Targeting Using Endogenous miRNA=== | ||
| + | |||
| + | Another application of our synthetic miRNA Kit profits of tissue specific endogenous miRNAs expression. These can be exploited for either Off- or On-Targeting. | ||
| + | To enable Off-Targeting, the GOI expressed on miTuner can be tagged with a miRNA binding site specific for one or a combination of endogenous miRNA of the tissue that is to be excluded from gene expression. | ||
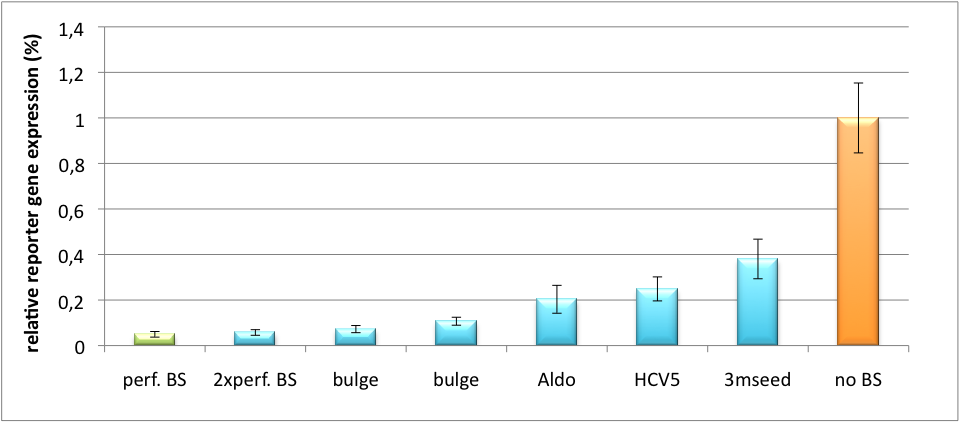
| + | In our experiment, we transfected Huh7 (human hepatoma) cells that endogenously overexpress miR122 with the miTuner construct after cloning different variations of binding sites for miR122 behind firefly luciferase. Figure 5 shows the results of the dual luciferase assay. Perfect binding sites result in almost complete inhibition of expression. | ||
| + | |||
| + | [[Image:HuH Offpng.png|thumb|center|500px|'''Figure 5: Knockdown of reporter gene expression due to endogenous miR122 that interferes with binding sites.''' Construct transfected to HuH cells to off-target those.]] | ||
| + | |||
| + | {| class="wikitable sortable" border="0" align="center" style="text-align: left" | ||
| + | |-bgcolor=#009be1 | ||
| + | |+ align="top, left"|'''Table 1: Mutated Binding Sites Against miR122''' | ||
| + | |Sequence||Mutation||Description | ||
| + | |- | ||
| + | |G ACAAACACCATTGTCACACTCCA TCTAGA GC||none||perfect BS | ||
| + | |- | ||
| + | |G ACAAACACCAT_ACGG_ACACTCCAGAGACACAAACACCAT_GAAG_ACACTCCA GC ||none||2x perfect BS | ||
| + | |- | ||
| + | |G C*C*CCTG*A*TGGGG*G*CGACACTCCA TCTAGA GC ||point mutations outside seed||HCV5 BS | ||
| + | |- | ||
| + | |TCGA G *AC*T*AA*GGCTGCT*CCAT*CAacactcca TCTAGA GC||one mutation inside seed||Aldo | ||
| + | |- | ||
| + | |TCGA G ACAAACACCATTGTCA*G*A*T*TC*G*A TCTAGA GC ||3 mutations in seed||3mutseed | ||
| + | |- | ||
| + | |G ACAAACACCAT_ACGA_ACACTCCA TCTAGA GC ||ACGA bulge||bulge region | ||
| + | |- | ||
| + | |TCGA G ACAAACACCAT_GCAG_ACACTCCA TCTAGA GC||GCAG bulge||bulge region | ||
| + | |} | ||
| + | |||
| + | ===On-Targeting Using Endogenous miRNA=== | ||
| + | |||
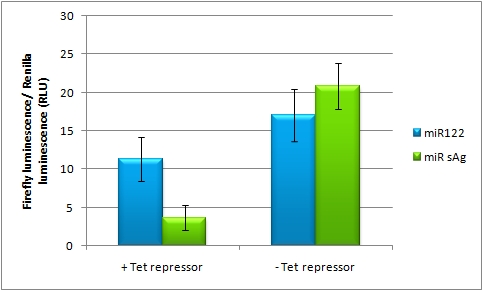
| + | In line with the Off-targeting approach, In the case of On-targeting the presence of a certain miRNA in a cell switches on expression of the GOI. This can be accomplished by using a repressor that is targeted by an endogenously expressed miRNA. We exemplified this scenario by using a Tet Repressor fused with a perfect binding site for miRNA 122, a liver-specific miRNA (Jopling et al., 2005). At the same time, the promoter expressing the GOI would be under control of a Tet Operator. Upon presence of the miRNA 122, the Tet Repressor would be knocked down, release the promoter and expression of the GOI could be established. | ||
| + | |||
| + | [[Image:101010on system.jpg|thumb|center|500px|'''Figure 6: Activation of gene expression upon Tet Repressor knockdown by liver-specific miR122]] | ||
| + | |||
| + | The miTuner Plasmid (driving the measurment BBa_K337038 luciferase from CMV_TetO2 promoter) was cotransfected with a TetR expression construct tagged with 4 perfect mir122 binding sites. Rescue of gene expression occurs in case of coexpression of shmir122. In the control experiment (coexpression of miRsAg) did not lead to Luc2 expression rescue, inicating that the on tuning is working. | ||
| + | |||
| + | |||
| + | [[Image:Ontuning_SV40_02.jpg|thumb|390px|left|Fig. 7: On-Tuning construct pBS_sv40_TetO2_Luc2 was cotransfected with a TetR tagged with 4 perfect miR122 binding sites and either an hcr (mir122 expression) construct or a mir155 control plasmid. Furthermore, a renilla construct was cotransfected also for normalization purposes. Transfection with hcr leads to higher Luc2 expression (rescue of expression) compared to the control, due to TetR knockdown. As positive control for the rescue, DOX was applied for avoid binding of the TetR to the Tet operator. ]] | ||
| + | [[Image:Offtuning_SV40.jpg|thumb|390px|right|Fig. 8: On-Tuning construct pBS_sv40_TetO2_Luc2 was cotransfected with a control TetR NOT tagged with mir122 binding sites and either an hcr (mir122 expression) construct or a mir155 control plasmid. Furthermore, a renilla construct was cotransfected also for normalization purposes. Transfection with hcr or mir122 lead to comparable expression ratios (NO rescue of expression via hac), indicating that the control TetR construct is not affected by either shRNA. As positive control for the rescue, DOX was applied again in order to avoid binding of the TetR to the Tet operator.]] | ||
| + | |||
| + | <br /><br /><br /><br /><br /><br /> | ||
| + | A pBS_SV40_TetO2_Luc2 construct was cotransfected with a Tet repressor construct tagged with 4 perfect mir122 binding sites. Fig. 7 shows a rescue of Luc2 expression in case of shmir122 expression (hcr construct), indicating, that the on-targeting is working. The right picture is the comparable control experiment using a not-binding site tagged TetR construct. As expected, no rescue of gene expression occurs in this control experiment (Fig. 8). | ||
| + | Those results indicate, that the on-tuning is working, in principle. In order to increase to rescue of gene expression, different TetR/pBS_SV40_TetO2_Luc2 ratios could be applied. | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
==Discussion== | ==Discussion== | ||
| + | <br /> | ||
| + | |||
| + | <!--Strikingly, the order of constructs in terms of knockdown for the imperfect binding sites is very similar in case of the tuning measurments with pBS_H1 and pBS_U6 shAAT. M4, M5 and M6 always show strong knockdown, whereas M9, M10 and M11 show only loose down-regulation. Consulting the binding site sequences, the weak knockdown can be addressed to bulges in the supplementary region or to complete lack of the 3' region of the binding site. Still high strength could be maintained due to only single nucleotide exchanges in the central region of the binding site. | ||
| + | The principle of smooth regulation was also demonstrated for miR122, a microRNA that is exclusively upregulated in hepatic cells. Referring binding sites were cloned into psiCHECK-2 backbone (Promega) and due to sequence mutations different Luciferase levels were detected again (Fig. 3).--> | ||
| + | <br /> | ||
| + | The On-construct measurments showed very promising results as well, having a clear firefly (Luc2) expression rescue in case the mir122 binding site TetR is cotransfected with the hcr (mir122) expression construct and not when being co-transfected with the control TetR (not binding site tagged). Therefor, the On-Tuning strategy is working. Adjustment of promoter strengths and ratios of TetR, hcr and the pBS_sv40_Tet02_Luc2 construct should be done in order to increase the rescue of gene expression compared to basal expression level of the Luc2 construct cotransfected with the TetR. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ==Application of miTuner== | ||
| + | |||
| + | === <i>In Vitro</i> Regulation of a Therapeutic Gene hAAT === | ||
| + | |||
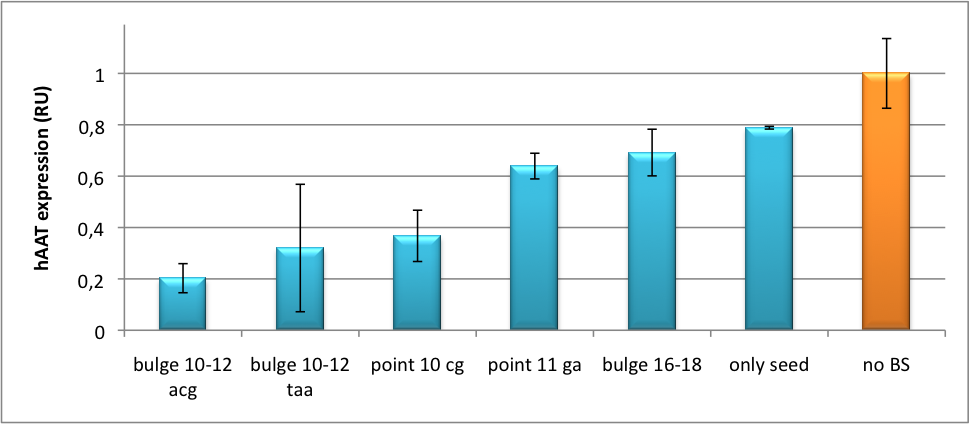
| + | We further tested our kit using a gene that is an interesting candidate for gene therapy, i. e. human alpha-1-antitrypsin (hAAT). Tight control of the genetic activity is fundamental, since deficiencies of hAAT can cause emphysema {{HDref|Lu et al., 2006}}. With our tuning kit we have a powerful mean at hand to mediate expression levels. In this approach, we tagged hAAT, that we used as our GOI, with binding sites (for miRsAg) that we measured and characterized with our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure miMeasure] construct beforehand (data not shown). There is some evidence, that the principle works also with this therapeutic gene in HeLa cells (fig. 7). This is a first potential therapeutic approach applying [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#ELISA ELISA] for measurements. | ||
| + | [[Image:HAAT ELISA.png|thumb|center|400px|'''Figure 7: hAAT expression in relative units depending on binding site properties.''' SV40 driven hAAT was fused to binding sites for miRsAg that was expressed from a co-transfected plasmid in HeLa cells.]] | ||
| + | It is obvious: different binding sites result in different knockdowns of gene expression. Some imperfect binding sites - e. g. single seed region - indicate even similar expression levels in accordance to the figures shown before. It can be stated, that the tuning idea seems to work for attempts varying in applied miRNAs, binding sites and reporter genes. | ||
| + | The hAAT as a GOI is worth testing because it is mainly secreted in liver -our target tissue of choice. Efficient transduction can be accomplished by infection with [https://2010.igem.org/Team:Heidelberg/Project/Capsid_Shuffling/Homology_Based selected viruses]. Dealing with hAAt intertwines our two approaches of specific gene therapy, therefor being a relevant field for future research. | ||
| + | |||
| + | ===<i>In Vivo</i> Validation=== | ||
| + | |||
| + | The constructs were tested in two different backbones: pBS_U6 and pBS_H1. Both are in viral context, meaning that they contain inverted terminal repeats (ITRs). The constructs can be packed into the capsid of an adeno-associated virus (AAV). Those constructs we also chose for [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Virus_Production virus production] to infect cells even more efficiently as compared to transfections. Because of the significant data, we decided to inject the viruses into mice to see the tuning effect also <i>[https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection in vivo]</i>. The pBS_H1 construct should be preferred for mice injections since the expressed synthetic shRNA miR against human alpha-1-antitrypsine (shhAAT) is cytotoxic in higher concentrations. The pBS_H1 backbone leads to moderate expression ranges, still obviously showing the tuning effect. | ||
| + | |||
| + | ===Modeling=== | ||
| + | |||
| + | After creating a binding site library and testing the miRNA-binding site interaction <i>in vitro</i>, we were able to compute an [https://2010.igem.org/Team:Heidelberg/Modeling/miGUI <i>in silico</i> model] based on a machine learning approach to predict knockdown efficiencies. A more detailed description of the different binding sites, we characterized can be found in our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure measurements] page. | ||
==Methods== | ==Methods== | ||
| - | |||
| - | + | ===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== | |
| + | The miTuner was [https://2010.igem.org/3A_Assembly assembled] out of different [https://2010.igem.org/Team:Heidelberg/Parts parts]. Cloning was done following [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Cloning standard protocols].<br> | ||
| + | |||
| + | Regulation of gene expression can be achieved by fusing miRNA binding sites right behind a GOI. In case a referring shRNA miR is expressed, the GOI is knocked down. Strength of regulation thereby depends on binding site properties. We are able to tune gene expression linearly over a broad range. This is a first proof of principle for various miRNA-mRNA interactions <i>in vitro</i>. Therefore, we transfected [https://2010.igem.org/Team:Heidelberg/Notebook/Material#Cell_lines HeLa cells] in principle with our [http://partsregistry.org/Part:BBa_K337036 pSMB_miTuner Plasmid HD3]. It turned out, that there was no obvious effect of different binding sites on reporter gene expression (data not shown). We assume that the RSV driving the shRNA miR is too weak for tight regulation of the referring binding site behind the GOI which is driven by the very strong CMV promoter. Only if a sufficient amount of shRNA miR binds to its target, translation is significantly repressed. Thus, we expressed the shRNA miR from a separate plasmid which was always co-transfected with the original tuning construct. The reporter genes - i. e. hFluc and hRluc - were also expressed from separate plasmids to get a reference as well as a transfection control. Then, we conducted a [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay Dual Luciferase Assay] for quantification of gene expression. | ||
| + | |||
| + | ===On- and Off-Targeting=== | ||
| + | |||
| + | Measurements were done in HeLa cells overexpressing miR122 from plasmid. Besides that, even endogenous miR122 levels were sufficient for off-targeting HuH cells (Fig. 4). A single perfect binding site leads to 95% knockdown, which seems to be maximum, since even a perfect binding site duplicate results in the same reporter gene expression. | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
==References== | ==References== | ||
| + | *Bartel DP: MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281-97.<br> | ||
| + | *Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005 Mar;3(3):e85.<br> | ||
| + | *Brown BD, Naldini L.: Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev *Genet. 2009 Aug;10(8):578-8<br> | ||
| + | *Chekulaeva M, Filipowicz W.:Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009 Jun;21(3):452-60.<br> | ||
| + | *Eulalio, A., Huntzinger, E., and Izaurralde, E. (2008). Getting to the root of miRNA-mediated gene silencing. Cell 132, 9-14. | ||
| + | *Bartel, D.P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281-297.<br> | ||
| + | *Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P.: Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005 Sep 2;309(5740):1577-81.<br> | ||
| - | |||
| - | + | <html> | |
| + | <div class="backtop"> | ||
| + | <a href="#top">↑</a> | ||
| + | </div> | ||
| + | </html> | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Bottom}} |
Latest revision as of 03:15, 28 October 2010

Synthetic miRNA Kit
miTuner - a kit for microRNA based gene expression tuning in mammalian cells
to plan, conduct and evaluate experiments dealing with miBricks (i. e. microRNA related Biobricks) as key regulators in mammalian cells.AbstractRegulation of any gene of interest has never been as easy as with our miRNA-based expression tuning kit miTuner. Rational design of synthetic miRNA binding sites according to our recommendations enables fine-tuning of gene expression in a range between 5% and 100%. Additionally, we offer Off- and On-targeting switches which effect GOI expression in only one or all but one tissue depending on endogenously occurring miRNAs. The tuning application is based on a dual promoter construct that expresses a GOI controlled by a synthetic miRNA which is expressed from either the same or a second construct (figure 1a). Differing miRNA-binding site interaction efficiencies caused by binding sites of different sequence properties are used to distinctly adjust expression strength of the GOI. For Off-targeting, the GOI is under control of miRNAs that are found in tissue where gene expression is thereupon silenced while the GOI can still be expressed in other tissues as visualized in figure 1b. On-targeting is based on the expression of the GOI from a promoter containing a Tet Operator (Tet02) that negatively regulates gene expression in the presence of a Tet Repressor (figure 1c). If the Tet Repressor tagged with binding sites for an endogenous miRNA, that is specifically expressed in the target cells/tissue. In consequence, the TetR is knocked down, releasing the promoter and enabling specific GOI expression. IntroductionMicroRNAs (miRNAs) are short endogenous, non-coding RNAs that mediate gene expression in a diversity of organisms [http://2010.igem.org/Team:Heidelberg/Project/miRNA_Kit#References (Bartel, 2004)]. Although the understanding of their biological functions is progressing remarkably, the exact mechanisms of regulation are still not unambiguously defined. However, it is commonly believed that miRNAs trigger target mRNA regulation by binding to 3’ untranslated region (UTR) of its target [http://2010.igem.org/Team:Heidelberg/Project/miRNA_Kit#References (Chekulaeva and Filipowicz, 2009)]. Exact principles of expression knockdown mediated by miRNA are still in debate [http://2010.igem.org/Team:Heidelberg/Project/miRNA_Kit#References (Eulalio et al., 2008)]. miTuner Kit componentsThe miTuner Kit consists of three basic components:
Please find basic information about the kit components and engineering of the kit here ResultsAll gene regulatory constructs for tuning, Off- or On-targeting can easily be assembled using BBB standard cloning from our miRNA Kit parts. After successful cloning, the constructs can be transfected onto a cell line of choice or transferred into a virus backbone for in vivo experiments. For our proof of principle, we used firefly luciferase normalized to Renilla luciferase on miTuner to characterize knockdown efficiencies of different binding sites and show Off- and On-targeting by mouse infection carried by an AAV virus. miTuner: Expression Fine-Tuning by Synthetic miRNAsThe data shows a precisely tuned expression from almost 0% to 100% (Fig. 2, Fig. 3). Lowest expression refers to complete knockdown through fusion of perfect binding sites (always green bar on the left hand side of the figures) to the reporter gene. Expression from a construct without binding sites is set as 100% (always orange column on the right hand side of the figures). In presence of the specific shRNA miR, gene expression was mediated to various levels through interactions with the different imperfect binding sites. Whereas, when an unspecific shRNA miR was expressed, gene expression remained unaffected (see raw data below). This reference shows that the binding sites were correctly designed, since they seem to interact specifically with a referring shRNA miR. Figure 2 shows the results of Dual-Luciferase measurements of the miTuner plasmid with binding sites against shhAAT behind firefly luciferase. The highest knockdown can be achieved by using a perfect binding site. Single mutations outside the seed region at position 11, 12 or 10-12 lead to knockdown between 10% and 60% compared to unregulated expression. Bulges close to the seed region or changes in the seed region itself lead to very low downregulation. Having only the seed region as a target for the miRNA also leads to a less efficient knockdown compared with binding sites containing flanking regions. 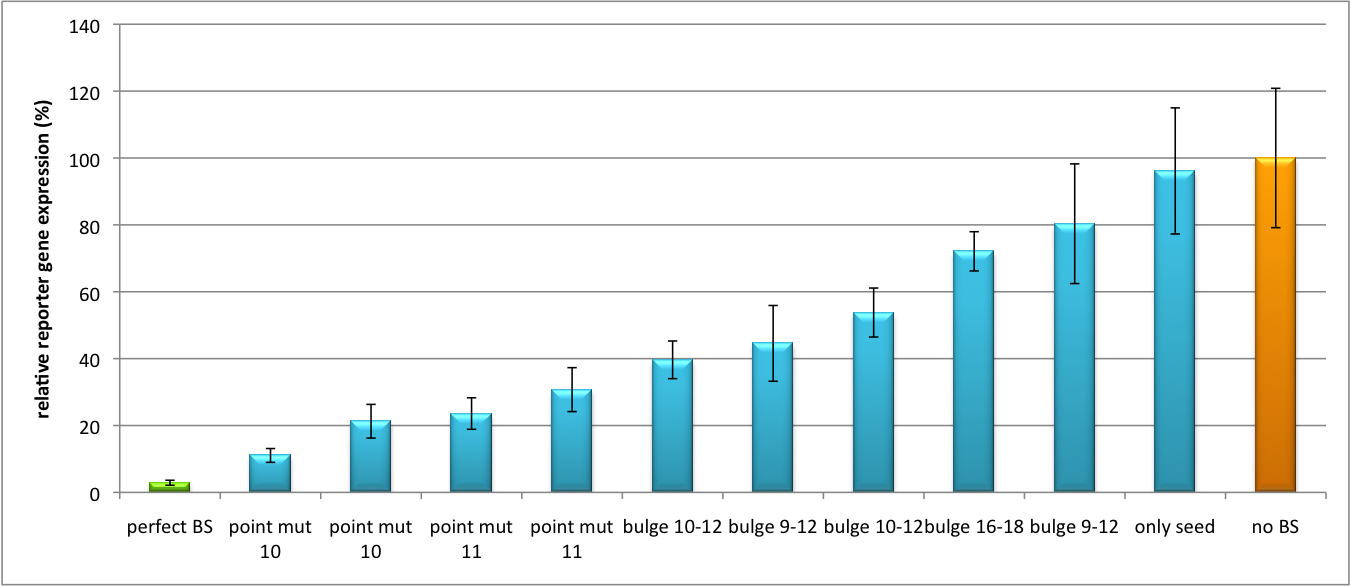 Figure 3: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_SV40_Luc2 construct cotransfected with a reference renilla construct. Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT. The shhAAT was expressed from a pBS_U6 plasmid Figure 3 shows the same assay using binding sites against shhAAT. This time, the shhAAT is driven by a U6 promoter, which is stronger than the H1 promoter used for driving the shRNA in the previous figure. The results are overall similar, with changes in or directly adjacent to the seed region having the highest impact on knockdown efficiency. We further analyzed binding sites derived from miR122 in the dual luciferase vector PsiCheck2 as can be seen in figure 4. Here we tested sixteen mutated binding sites in order to observe minute fine-tuning between one binding site and the next. Mutated Binding sites 123, 133, 134, 135 and 158 contain 4bp-bulges (non-paired regions) that don not seem to diminish knockdown efficiency much. 107 contains one binding site, while 134 and 135 contain two binding sites for the same miRNA and show a stronger knockdown than 107. Off-Targeting Using Endogenous miRNAAnother application of our synthetic miRNA Kit profits of tissue specific endogenous miRNAs expression. These can be exploited for either Off- or On-Targeting. To enable Off-Targeting, the GOI expressed on miTuner can be tagged with a miRNA binding site specific for one or a combination of endogenous miRNA of the tissue that is to be excluded from gene expression. In our experiment, we transfected Huh7 (human hepatoma) cells that endogenously overexpress miR122 with the miTuner construct after cloning different variations of binding sites for miR122 behind firefly luciferase. Figure 5 shows the results of the dual luciferase assay. Perfect binding sites result in almost complete inhibition of expression.
On-Targeting Using Endogenous miRNAIn line with the Off-targeting approach, In the case of On-targeting the presence of a certain miRNA in a cell switches on expression of the GOI. This can be accomplished by using a repressor that is targeted by an endogenously expressed miRNA. We exemplified this scenario by using a Tet Repressor fused with a perfect binding site for miRNA 122, a liver-specific miRNA (Jopling et al., 2005). At the same time, the promoter expressing the GOI would be under control of a Tet Operator. Upon presence of the miRNA 122, the Tet Repressor would be knocked down, release the promoter and expression of the GOI could be established. The miTuner Plasmid (driving the measurment BBa_K337038 luciferase from CMV_TetO2 promoter) was cotransfected with a TetR expression construct tagged with 4 perfect mir122 binding sites. Rescue of gene expression occurs in case of coexpression of shmir122. In the control experiment (coexpression of miRsAg) did not lead to Luc2 expression rescue, inicating that the on tuning is working.
 Fig. 7: On-Tuning construct pBS_sv40_TetO2_Luc2 was cotransfected with a TetR tagged with 4 perfect miR122 binding sites and either an hcr (mir122 expression) construct or a mir155 control plasmid. Furthermore, a renilla construct was cotransfected also for normalization purposes. Transfection with hcr leads to higher Luc2 expression (rescue of expression) compared to the control, due to TetR knockdown. As positive control for the rescue, DOX was applied for avoid binding of the TetR to the Tet operator.  Fig. 8: On-Tuning construct pBS_sv40_TetO2_Luc2 was cotransfected with a control TetR NOT tagged with mir122 binding sites and either an hcr (mir122 expression) construct or a mir155 control plasmid. Furthermore, a renilla construct was cotransfected also for normalization purposes. Transfection with hcr or mir122 lead to comparable expression ratios (NO rescue of expression via hac), indicating that the control TetR construct is not affected by either shRNA. As positive control for the rescue, DOX was applied again in order to avoid binding of the TetR to the Tet operator.
Discussion
Application of miTunerIn Vitro Regulation of a Therapeutic Gene hAATWe further tested our kit using a gene that is an interesting candidate for gene therapy, i. e. human alpha-1-antitrypsin (hAAT). Tight control of the genetic activity is fundamental, since deficiencies of hAAT can cause emphysema [http://2010.igem.org/Team:Heidelberg/Project/miRNA_Kit#References (Lu et al., 2006)]. With our tuning kit we have a powerful mean at hand to mediate expression levels. In this approach, we tagged hAAT, that we used as our GOI, with binding sites (for miRsAg) that we measured and characterized with our miMeasure construct beforehand (data not shown). There is some evidence, that the principle works also with this therapeutic gene in HeLa cells (fig. 7). This is a first potential therapeutic approach applying ELISA for measurements. It is obvious: different binding sites result in different knockdowns of gene expression. Some imperfect binding sites - e. g. single seed region - indicate even similar expression levels in accordance to the figures shown before. It can be stated, that the tuning idea seems to work for attempts varying in applied miRNAs, binding sites and reporter genes. The hAAT as a GOI is worth testing because it is mainly secreted in liver -our target tissue of choice. Efficient transduction can be accomplished by infection with selected viruses. Dealing with hAAt intertwines our two approaches of specific gene therapy, therefor being a relevant field for future research. In Vivo ValidationThe constructs were tested in two different backbones: pBS_U6 and pBS_H1. Both are in viral context, meaning that they contain inverted terminal repeats (ITRs). The constructs can be packed into the capsid of an adeno-associated virus (AAV). Those constructs we also chose for virus production to infect cells even more efficiently as compared to transfections. Because of the significant data, we decided to inject the viruses into mice to see the tuning effect also in vivo. The pBS_H1 construct should be preferred for mice injections since the expressed synthetic shRNA miR against human alpha-1-antitrypsine (shhAAT) is cytotoxic in higher concentrations. The pBS_H1 backbone leads to moderate expression ranges, still obviously showing the tuning effect. ModelingAfter creating a binding site library and testing the miRNA-binding site interaction in vitro, we were able to compute an in silico model based on a machine learning approach to predict knockdown efficiencies. A more detailed description of the different binding sites, we characterized can be found in our measurements page. MethodsmiTuner: Expression Fine-Tuning by Synthetic miRNAsThe miTuner was assembled out of different parts. Cloning was done following standard protocols. Regulation of gene expression can be achieved by fusing miRNA binding sites right behind a GOI. In case a referring shRNA miR is expressed, the GOI is knocked down. Strength of regulation thereby depends on binding site properties. We are able to tune gene expression linearly over a broad range. This is a first proof of principle for various miRNA-mRNA interactions in vitro. Therefore, we transfected HeLa cells in principle with our [http://partsregistry.org/Part:BBa_K337036 pSMB_miTuner Plasmid HD3]. It turned out, that there was no obvious effect of different binding sites on reporter gene expression (data not shown). We assume that the RSV driving the shRNA miR is too weak for tight regulation of the referring binding site behind the GOI which is driven by the very strong CMV promoter. Only if a sufficient amount of shRNA miR binds to its target, translation is significantly repressed. Thus, we expressed the shRNA miR from a separate plasmid which was always co-transfected with the original tuning construct. The reporter genes - i. e. hFluc and hRluc - were also expressed from separate plasmids to get a reference as well as a transfection control. Then, we conducted a Dual Luciferase Assay for quantification of gene expression. On- and Off-TargetingMeasurements were done in HeLa cells overexpressing miR122 from plasmid. Besides that, even endogenous miR122 levels were sufficient for off-targeting HuH cells (Fig. 4). A single perfect binding site leads to 95% knockdown, which seems to be maximum, since even a perfect binding site duplicate results in the same reporter gene expression.
References
|
||||||||||||||||||||||||||
 "
"