Team:Northwestern/Project
From 2010.igem.org
(Difference between revisions)
| (13 intermediate revisions not shown) | |||
| Line 129: | Line 129: | ||
{| align="center" border="0" width="75%" cellpadding="20" | {| align="center" border="0" width="75%" cellpadding="20" | ||
| - | |colspan="2"| | + | |colspan="2" style="padding:0"| |
<html> | <html> | ||
<table align="center"> | <table align="center"> | ||
| Line 150: | Line 150: | ||
|- | |- | ||
|align="left" valign="top" width="80%" cellpadding="10"| | |align="left" valign="top" width="80%" cellpadding="10"| | ||
| + | =='''Overview'''== | ||
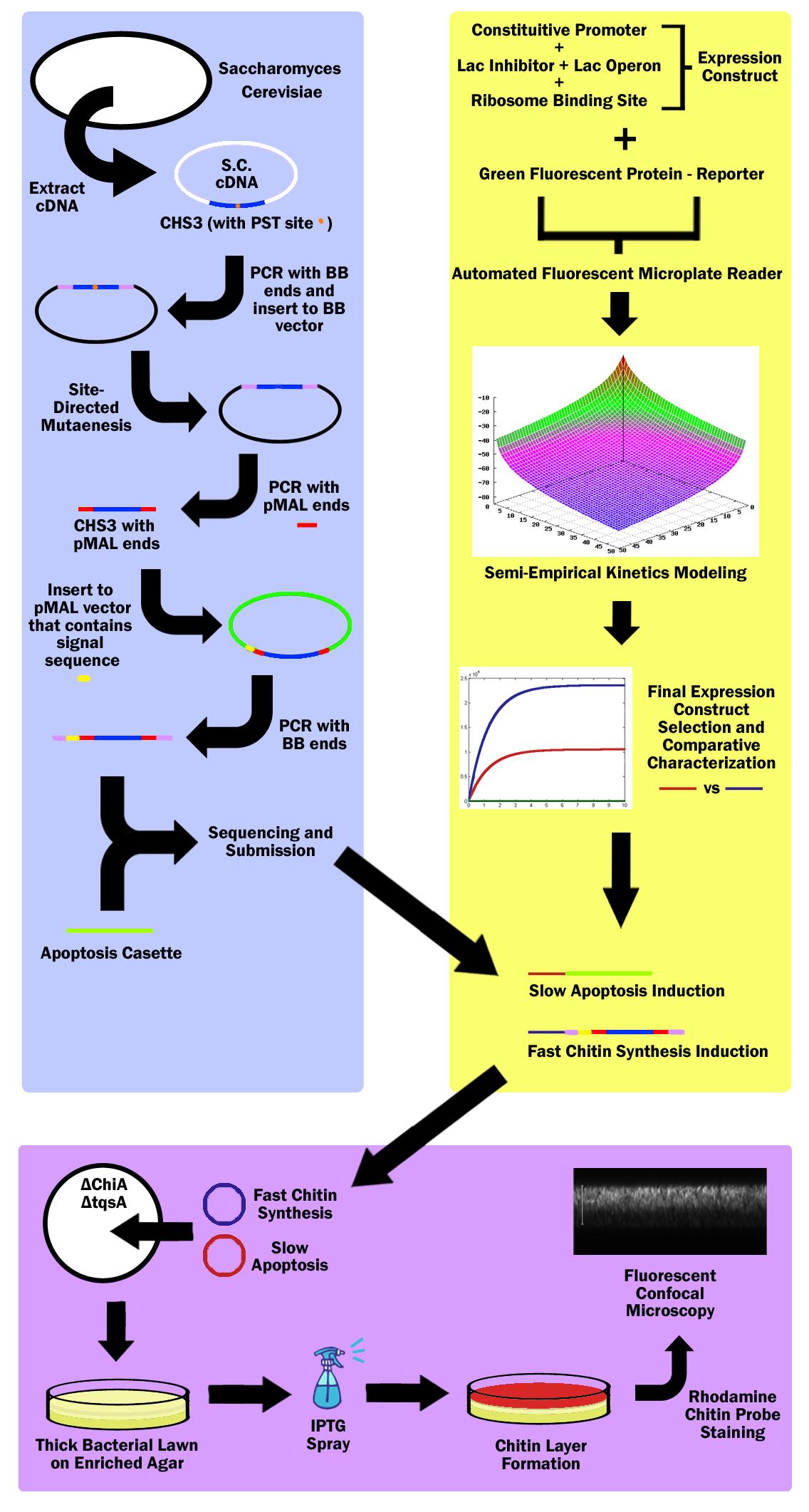
| - | Our main project is to simulate mammalian skin using chitin in E. Coli | + | Our main project is to simulate mammalian skin using chitin in E. Coli. |
| - | |||
| - | + | In mammalian skin, mitosis occurs in the basal layer of the epithelial cells and cells travel outwards towards the surface of the skin as they mature. As the cells move further away from the basal layer, they die and their cytoplasm is released and the cells are filled with keratin, thus forming a continuously regenerating protective layer on the outer-most part of epithelial layer; our project is modeled after this. | |
| - | |||
| - | + | First, a specific strain of Escherichia Coli is used to generate a thick bacterial biofilm lawn, which is formed on an enriched LB agar plate. The strain also does not produce chitinase so that the produced chitin is not digested. | |
| - | |||
| + | Then a fast, high-expression signalling pathway initiates production of chitin in the top-most layer of the lawn. The gene for chitin synthesis is extracted from the genome of Saccharomyces Cerevisiae, more commonly known as Baker's Yeast, while the signalling pathway and expression parts are identified from the parts registry and extracted from the iGEM 2010 Distribution kit. | ||
| - | |||
| - | + | Simultaneously, a slow, medium-expression signalling pathway initiates production of a cell lysis expression vector in the top-most layer of the lawn. The apoptosis part used is the T4 Bacteriophage apoptosis cassette, and this part as well as the signalling and expression parts, are identified from the parts registry and extracted from the iGEM 2010 Distribution kit. | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | With both vectors, the E.Coli cells in the top layer generates chitin then lyses, thus depositing a layer of chitin in the top-most layer of the bacterial lawn. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | + | To identify the expression of chitin, a chitin-specific rhodamine stain is used. To identify the efficacy of the apoptosis cassette, a live-dead assay is used. With these stains, the colony can be imaged vertically through confocal microscopy to show the deposition of Chitin in the top layer. | |
| - | + | ||
| + | Whenever there is damage to the chitin layer, the signalling pathway can be reinitiated to regenerate chitin in the damaged location. | ||
| + | |||
| + | |||
| + | ||[[Image:PosterChild.jpg|400px|center]] | ||
| + | |- | ||
| + | |colspan="2" cellpadding="10"| | ||
==Strategy== | ==Strategy== | ||
| Line 213: | Line 198: | ||
===MAGE=== | ===MAGE=== | ||
| + | The Multiplex Automated Genome Engineering (MAGE) protocol developed by H. H. Wang. et al. is used to acquire double knockouts. The MAGE protocol is an accelerated evolution procedure where oligonucleotides are electroporated into the cells in order to introduce mutations in the genome during replication by binding as lagging strand primers, taking advantage of the lambda-red construct. | ||
Latest revision as of 23:58, 23 October 2010
| Home | Brainstorm | Team | Acknowledgements | Project | Human Practices | Parts | Notebook | Calendar | Protocol | Safety | Links | References | Media | Contact |
|---|
 "
"