Team:USTC/Project/overview
From 2010.igem.org
| (12 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<html> | <html> | ||
| + | <style> | ||
| + | #transparent{ | ||
| + | position:relative; | ||
| + | left:255px; | ||
| + | width:675px; | ||
| + | height:3000px; | ||
| + | padding:10px; | ||
| + | background-image:url(https://static.igem.org/mediawiki/2010/a/a0/Transppa.png); | ||
| + | background-repeat:repeat; | ||
| + | filter:alpha(opacity=90); | ||
| + | opacity:0.90; | ||
| + | border-bottom-right-radius:20px; | ||
| + | border-bottom-left-radius:20px; | ||
| + | border-top-left-radius:20px; | ||
| + | border-top-right-radius:20px; | ||
| + | } | ||
| + | </style> | ||
| + | <div id="transparent"> | ||
| + | </html> | ||
| - | < | + | <html> |
| - | + | <style> | |
| - | <style | + | .firstletter{ |
| - | + | font-size:3em; | |
| - | + | line-height:0.95; | |
| + | float:left; | ||
</style> | </style> | ||
| - | </ | + | </html> |
| + | <p ><span class="firstletter">C</span> | ||
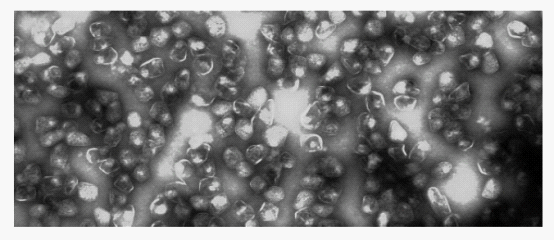
| + | ompartmentalization is a smart strategy for offering the segregated and characterasitic microenvironments for diverse metabolic activities in living creature. The organelles in eucaryotic cell provide a stong illustration. Meanwhile, some bacteria contain microcompartments consisting of a large virion-like protein shell and encapsulated sequentially acting enzymes. According to the different metabolic pathway carrying out by enzymes, microcompartments are divided into various types, including carboxysome, propanediol utilization compartments (Pdu) and ethalnolamine utilization compartment (Eut) etc.. Carboxysome, isolated from the chemoautotroph Halothiobacillus neapolitanus, consisted mainly of the CO2 - fixing enzyme ribulose bisphosphate carboxylase/oxygenase (RuBisCO), play a role in CO2-fixation. Eut Microcompartments (Eut), containing unknown enzyme, convert ethanolamine to acetaldehyde and then to acetyl-CoA before the acetaldehyde can escape into the cytosol. Pdu Microcompartments (Pdu)(figure1), degrade 1,2-PD to propionaldehyde by an adenosylcobalamin-dependent diol dehydratase, followed by the conversion of propionaldehyde to propionyl-CoA and 1-propanol without escape of the aldehyde into the cytosol. An article revealed that a short N-terminal peptide is necessary and sufficient for packaging enzymes into the lumen of an Pdu microcompartment, so we finally focus on the Pdu microcompartment.</p> | ||
| - | + | [[Image:Ustc2010-dd-background1.png ]] | |
| - | + | Fig 1: Purified Pdu microcompartment | |
| - | + | ||
| - | + | In 1994, genetic studies indicated that a BMC-domain protein was involved in B 12-dependent 1,2-propanediol degradation by S.enterica. A few years later, electron microscopy showed that S.enterica conditionally formed microcompartments during B12-dependent growth on 1,2-propanediol and further studies established that microcompartments function in this process. Then the Pdu microcompartments is officially accepted by scientists. The Pdu microcompartments are very large multiprotein complexes and 100-150 nm in cross section with a 3-4 nm protein shell. Their shape is roughly polyhedral, but they are more irregular than carboxysomes which is nearly a icosahedron. | |
| + | |||
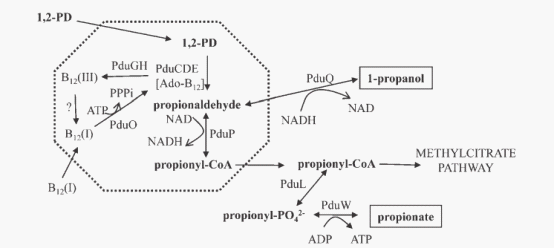
| + | The current model(figure 2) of the Pdu microcompartment proposes that the first two steps of 1,2-propanediol degradation are confined to the lumen of a microcompartment so that propionaldehyde can be sequestered. 1,2-propanediol travels through the microcompartment shell into the lumen and simultaneously converting to propionaldehyde by B12 -dependent diol dehydratase. Propionaldehyde dehydrogenase then catalyzes the conversion: propionaldehyde + HS-CoA +NAD+→propionyl-CoA + NADH + H+. Propionyl-CoA exits the microcompartment and continues through the 1,2-propanediol degradative pathway to propionate and 1-propanol, or enters the methylcitrate pathway where it is converted to pyruvate and succinate. These processes allow S. enterica to grow efficiently on 1,2-propanediol as a sole carbon and energy source while at the same time isolating the cytotoxic propionaldehyde. | ||
| - | + | [[Image:Ustc2010-dd-bg2.png ]] | |
| + | Fig 2: Model for 1,2-propanediol degradation | ||
| + | |||
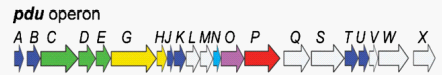
| + | The protein content of the Pdu microcompartment supports the functional model described above. The Pdu microcompartments were purified intact (figure 1). They consist of 14 different major polypeptides (PduABB’CDEGHJKOPTU).(figure 3). Different proteins have diverse function among the complex. For example, PduCDE is enzyme of the Pdu microcomparmet, B12-dependent diol dehydratase. And PduABB0JKTUF make up of the ideal shell of the microcompartment, More detailed information can be gotten from the notation of figure 3 and reference. | ||
| + | |||
| + | [[Image:Ustc2010-dd-bg3.png]] | ||
| + | Fig 3: Gene organization for the Pdu operon | ||
| + | PduABB'JKTU : shell of pdu microcompartment | ||
| + | PduGH, PduO, PduS :three enzymes for B 12 metabolism | ||
| + | PduX :one enzyme used for the de novo synthesis of B 12 | ||
| + | PduCDE, PduL, PduP, PduQ, PduW : five catabolic enzymes that mediate 1,2-propanediol Degradation | ||
| + | PduN : a protein with homology to the pentamer proposed to form the carboxysome vertices | ||
| + | |||
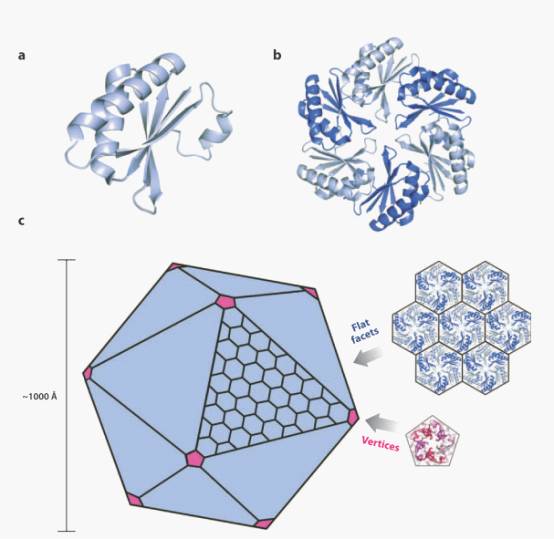
| + | Although three kinds of bacterial compartments provide diverse metabolic functions, they share an evolutionarily related shell, which is defined by a conserved protein domain, BMC protein domain. The BMC protein monomers(figure4 a) adopts an α/β fold with a central four-stranded anti-parallel sheet flanked by small helices. And extensive interactions hold the monomers together within a hexamer (figure4 b), Hexameric building blocks of the BMC proteins can assemble into a molecular layer which forms flat facets of the polyhedral shells (figure 4c up). The pentameric proteins from the carboxysome (figure4c down) have been argued to form vertices of the icosahedral carboxysome (left) . The Pdu microcompartments are less geometrically regular than the carboxysome and are potentially more complex. | ||
| + | |||
| + | [[Image:Ustc2010-dd-bg4.png ]] | ||
| + | Fig 4: Ideal model for assembly of bacterial microcompartment | ||
| + | |||
| + | Reference | ||
| + | |||
| + | ---- | ||
| + | 1. CG Fan, SQ Cheng, Y Liu, CM Escobar, CS Crowley, RE Jefferson, TO Yeates, TA Bobik: Short N-terminal sequences package proteins into bacterial microcompartments. Proceedings of the National Academy of Sciences of the United States of America 2010, 107:7509-7514. | ||
| + | |||
| + | 2. Tsai Y, Sawaya MR, Yeates TO. 2009. Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallogr. D 65:980–88 | ||
| + | |||
| + | 3. Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, et al. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–38 | ||
| + | |||
| + | 4. Havemann GD, Bobik TA. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol 185:5086–5095. | ||
Latest revision as of 22:10, 27 October 2010
C ompartmentalization is a smart strategy for offering the segregated and characterasitic microenvironments for diverse metabolic activities in living creature. The organelles in eucaryotic cell provide a stong illustration. Meanwhile, some bacteria contain microcompartments consisting of a large virion-like protein shell and encapsulated sequentially acting enzymes. According to the different metabolic pathway carrying out by enzymes, microcompartments are divided into various types, including carboxysome, propanediol utilization compartments (Pdu) and ethalnolamine utilization compartment (Eut) etc.. Carboxysome, isolated from the chemoautotroph Halothiobacillus neapolitanus, consisted mainly of the CO2 - fixing enzyme ribulose bisphosphate carboxylase/oxygenase (RuBisCO), play a role in CO2-fixation. Eut Microcompartments (Eut), containing unknown enzyme, convert ethanolamine to acetaldehyde and then to acetyl-CoA before the acetaldehyde can escape into the cytosol. Pdu Microcompartments (Pdu)(figure1), degrade 1,2-PD to propionaldehyde by an adenosylcobalamin-dependent diol dehydratase, followed by the conversion of propionaldehyde to propionyl-CoA and 1-propanol without escape of the aldehyde into the cytosol. An article revealed that a short N-terminal peptide is necessary and sufficient for packaging enzymes into the lumen of an Pdu microcompartment, so we finally focus on the Pdu microcompartment.
Fig 1: Purified Pdu microcompartment
In 1994, genetic studies indicated that a BMC-domain protein was involved in B 12-dependent 1,2-propanediol degradation by S.enterica. A few years later, electron microscopy showed that S.enterica conditionally formed microcompartments during B12-dependent growth on 1,2-propanediol and further studies established that microcompartments function in this process. Then the Pdu microcompartments is officially accepted by scientists. The Pdu microcompartments are very large multiprotein complexes and 100-150 nm in cross section with a 3-4 nm protein shell. Their shape is roughly polyhedral, but they are more irregular than carboxysomes which is nearly a icosahedron.
The current model(figure 2) of the Pdu microcompartment proposes that the first two steps of 1,2-propanediol degradation are confined to the lumen of a microcompartment so that propionaldehyde can be sequestered. 1,2-propanediol travels through the microcompartment shell into the lumen and simultaneously converting to propionaldehyde by B12 -dependent diol dehydratase. Propionaldehyde dehydrogenase then catalyzes the conversion: propionaldehyde + HS-CoA +NAD+→propionyl-CoA + NADH + H+. Propionyl-CoA exits the microcompartment and continues through the 1,2-propanediol degradative pathway to propionate and 1-propanol, or enters the methylcitrate pathway where it is converted to pyruvate and succinate. These processes allow S. enterica to grow efficiently on 1,2-propanediol as a sole carbon and energy source while at the same time isolating the cytotoxic propionaldehyde.
Fig 2: Model for 1,2-propanediol degradation
The protein content of the Pdu microcompartment supports the functional model described above. The Pdu microcompartments were purified intact (figure 1). They consist of 14 different major polypeptides (PduABB’CDEGHJKOPTU).(figure 3). Different proteins have diverse function among the complex. For example, PduCDE is enzyme of the Pdu microcomparmet, B12-dependent diol dehydratase. And PduABB0JKTUF make up of the ideal shell of the microcompartment, More detailed information can be gotten from the notation of figure 3 and reference.
Fig 3: Gene organization for the Pdu operon
PduABB'JKTU : shell of pdu microcompartment
PduGH, PduO, PduS :three enzymes for B 12 metabolism
PduX :one enzyme used for the de novo synthesis of B 12
PduCDE, PduL, PduP, PduQ, PduW : five catabolic enzymes that mediate 1,2-propanediol Degradation
PduN : a protein with homology to the pentamer proposed to form the carboxysome vertices
Although three kinds of bacterial compartments provide diverse metabolic functions, they share an evolutionarily related shell, which is defined by a conserved protein domain, BMC protein domain. The BMC protein monomers(figure4 a) adopts an α/β fold with a central four-stranded anti-parallel sheet flanked by small helices. And extensive interactions hold the monomers together within a hexamer (figure4 b), Hexameric building blocks of the BMC proteins can assemble into a molecular layer which forms flat facets of the polyhedral shells (figure 4c up). The pentameric proteins from the carboxysome (figure4c down) have been argued to form vertices of the icosahedral carboxysome (left) . The Pdu microcompartments are less geometrically regular than the carboxysome and are potentially more complex.
Fig 4: Ideal model for assembly of bacterial microcompartment
Reference
1. CG Fan, SQ Cheng, Y Liu, CM Escobar, CS Crowley, RE Jefferson, TO Yeates, TA Bobik: Short N-terminal sequences package proteins into bacterial microcompartments. Proceedings of the National Academy of Sciences of the United States of America 2010, 107:7509-7514.
2. Tsai Y, Sawaya MR, Yeates TO. 2009. Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallogr. D 65:980–88
3. Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, et al. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–38
4. Havemann GD, Bobik TA. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol 185:5086–5095.
 "
"