|
|
| (11 intermediate revisions not shown) |
| Line 39: |
Line 39: |
| | After parts have been moved to the shipping plasmid and checked by agarose gel electrophoresis. | | After parts have been moved to the shipping plasmid and checked by agarose gel electrophoresis. |
| | | | |
| - | The parts we sent to MIT are: | + | The parts we sent to MIT and submitted to the registry are: |
| | | | |
| | <table border='1' align='center'> | | <table border='1' align='center'> |
| Line 75: |
Line 75: |
| | | | |
| | | | |
| - | The parts we submitted to the Registry are: | + | The parts we have in freezer and we only submitted to the Registry are: |
| | <table border='1' align='center'> | | <table border='1' align='center'> |
| | <tr><th>Part name</th><th>Wiki name</th><th>Plasmid</th></tr> | | <tr><th>Part name</th><th>Wiki name</th><th>Plasmid</th></tr> |
| - | <tr><td><partinfo>BBa_K300019</partinfo></td><td>I12</td><td><partinfo>BBa_J61002</partinfo></td></tr>
| |
| | <tr><td><partinfo>BBa_K300021</partinfo></td><td>I8</td><td><partinfo>BBa_J61002</partinfo></td></tr> | | <tr><td><partinfo>BBa_K300021</partinfo></td><td>I8</td><td><partinfo>BBa_J61002</partinfo></td></tr> |
| | <tr><td><partinfo>BBa_K300022</partinfo></td><td>I9</td><td><partinfo>BBa_J61002</partinfo></td></tr> | | <tr><td><partinfo>BBa_K300022</partinfo></td><td>I9</td><td><partinfo>BBa_J61002</partinfo></td></tr> |
| | <tr><td><partinfo>BBa_K300023</partinfo></td><td>I10</td><td><partinfo>BBa_J61002</partinfo></td></tr> | | <tr><td><partinfo>BBa_K300023</partinfo></td><td>I10</td><td><partinfo>BBa_J61002</partinfo></td></tr> |
| - | <tr><td><partinfo>BBa_K300024</partinfo></td><td>I14</td><td><partinfo>BBa_J61002</partinfo></td></tr> | + | <tr><td><partinfo>BBa_K300091</partinfo></td><td>I63</td><td><partinfo>pSB1AK3</partinfo></td></tr> |
| - | <tr><td><partinfo></partinfo></td><td></td><td><partinfo></partinfo></td></tr> | + | <tr><td><partinfo>BBa_K300092</partinfo></td><td>I64</td><td><partinfo>pSB1A2</partinfo></td></tr> |
| | + | <tr><td><partinfo>BBa_K300099</partinfo></td><td>I65</td><td><partinfo>pSB1A2</partinfo></td></tr> |
| | + | <tr><td><partinfo>BBa_K300980</partinfo></td><td>YEAST MR.G</td><td>pMA commercial vector</td></tr> |
| | + | <tr><td><partinfo>BBa_K300982</partinfo></td><td>CREAM</td><td>-</td></tr> |
| | + | <tr><td><partinfo>BBa_K300983</partinfo></td><td>CRIM MR.G</td><td>pMK-RQ commercial vector</td></tr> |
| | + | <!--tr><td><partinfo></partinfo></td><td></td><td><partinfo></partinfo></td></tr--> |
| | + | </table> |
| | | | |
| | | | |
| | + | ---- |
| | | | |
| - | <tr><td><partinfo></partinfo></td><td></td><td><partinfo></partinfo></td></tr>
| |
| | | | |
| - | </table> | + | <div align="right"><small>[[#indice|^top]]</small></div> |
| | | | |
| | + | ==October, 19th== |
| | + | <b>Wiki update and definition of results.</b> |
| | | | |
| | ---- | | ---- |
| - | In order to obtain better results with screening PCR for the E. coli genome integration project, we perform amplifications using an annealing temperature of 63°C.
| + | Every clones of E. coli genome integration project were amplified with PCR to verify their lengths and their positions into the genome. |
| - | Screening PCR was performed on MC42(A-B-C), MC43(A-B-C), MG42(B-C), MG43(A-B-C), MC1061 and MG1655. Primers used were BBa_K300975-BBa_K300976, BBa_K300976-BBa_K300977, BBa_K300977-BBa_K300978. | + | In order to obtain better results we perform all the amplifications using an annealing temperature of 63°C. |
| | + | Screening PCR was performed on MC42(A-B-C), MC43(A-B-C), MG42(B-C), MG43(A-B-C), MC1061 and MG1655. Primers used were BBa_K300975-BBa_K300976 (P1-P2), BBa_K300976-BBa_K300977 (P2-P3), BBa_K300977-BBa_K300978 (P3-P4). |
| | | | |
| - | [[Image:UNIPV10_19-10-10_MC42ABC_MC43ABC_MG42BC_MG43ABC_BLANK_MC1061_MG1655(TUTTO_P1_P2).jpg|thumb|300px|center|The order of the samples was the follow: MC42(A,B,C), MC43(A,B,C), MG42(B,C), MG43(A,B,C), blank, MC1061, MG1655. Primers were BBa_K300975-BBa_K300976.]] | + | [[Image:UNIPV10_19_10_10_MC42ABC_MC43ABC_MG42BC_MG43ABC_BLANK_MC1061_MG1655(TUTTI P1-P2).jpg|thumb|300px|center|The order of the samples was the follow: MC42(A,B,C), MC43(A,B,C), MG42(B,C), MG43(A,B,C), blank, MC1061, MG1655. Primers were BBa_K300975-BBa_K300976 (P1-P2).]] |
| | + | |
| | + | [[Image:UNIPV10_19_10_10_MC42ABC_MC43ABC_MG42BC_MG43ABC_BLANK_MC1061_MG1655(TUTTI P2-P3).jpg|thumb|300px|center|The order of the samples was the follow: MC42(A,B,C), MC43(A,B,C), MG42(B,C), MG43(A,B,C), blank, MC1061, MG1655. Primers were BBa_K300976-BBa_K300977 (P2-P3).]] |
| | + | |
| | + | [[Image:UNIPV10_19_10_10_MC42ABC_MC43ABC_MG42BC_MG43ABC_BLANK_MC1061_MG1655(TUTTI P3-P4).jpg|thumb|300px|center|The order of the samples was the follow: MC42(A,B,C), MC43(A,B,C), MG42(B,C), MG43(A,B,C), blank, MC1061, MG1655. Primers were BBa_K300977-BBa_K300978 (P3-P4).]] |
| | | | |
| | | | |
| | | | |
| - | <div align="right"><small>[[#indice|^top]]</small></div>
| |
| | | | |
| - | ==October, 19th==
| |
| - | <b>Wiki update and definition of results.</b>
| |
| | <div align="right"><small>[[#indice|^top]]</small></div> | | <div align="right"><small>[[#indice|^top]]</small></div> |
| | | | |
|
|
|
|
|
OCTOBER: WEEK 3
October, 18th
We have finally completed the parts transfer to <partinfo>pSB1C3</partinfo> plasmid.
All the parts that will be sent to the registry were previously screened by UNIPV-Pavia TEAM, as reported in this notebook and all of them were sequenced and sequencing results were correct.
After parts have been moved to the shipping plasmid and checked by agarose gel electrophoresis.
The parts we sent to MIT and submitted to the registry are:
| Part name | Wiki name | Plasmid | Number |
|---|
| <partinfo>BBa_K300005</partinfo> | I0 | pSB1C3 | 1 |
| <partinfo>BBa_K300006</partinfo> | I1 | pSB1C3 | 2 |
| <partinfo>BBa_K300009</partinfo> | I3 | pSB1C3 | 3 |
| <partinfo>BBa_K300008</partinfo> | I5 | pSB1C3 | 4 |
| <partinfo>BBa_K300024</partinfo> | I7 | pSB1C3 | 5 |
| <partinfo>BBa_K300019</partinfo> | I12 | pSB1C3 | 6 |
| <partinfo>BBa_K300030</partinfo> | I14 | pSB1C3 | 7 |
| <partinfo>BBa_K300028</partinfo> | I15 | pSB1C3 | 8 |
| <partinfo>BBa_K300029</partinfo> | I16 | pSB1C3 | 9 |
| <partinfo>BBa_K300025</partinfo> | I17 | pSB1C3 | 10 |
| <partinfo>BBa_K300026</partinfo> | I18 | pSB1C3 | 11 |
| <partinfo>BBa_K300027</partinfo> | I19 | pSB1C3 | 12 |
| <partinfo>BBa_K300002</partinfo> | I20 | pSB1C3 | 13 |
| <partinfo>BBa_K300003</partinfo> | I21 | pSB1C3 | 14 |
| <partinfo>BBa_K300031</partinfo> | I22 | pSB1C3 | 15 |
| <partinfo>BBa_K300081</partinfo> | I26 | pSB1C3 | 16 |
| <partinfo>BBa_K300079</partinfo> | I31 | pSB1C3 | 17 |
| <partinfo>BBa_K300080</partinfo> | I35 | pSB1C3 | 18 |
| <partinfo>BBa_K300086</partinfo> | I47 | pSB1C3 | 19 |
| <partinfo>BBa_K300088</partinfo> | I48 | pSB1C3 | 20 |
| <partinfo>BBa_K300090</partinfo> | I49 | pSB1C3 | 21 |
| <partinfo>BBa_K300082</partinfo> | I55 | pSB1C3 | 22 |
| <partinfo>BBa_K300084</partinfo> | I78 | pSB1C3 | 23 |
| <partinfo>BBa_K300083</partinfo> | I79 | pSB1C3 | 24 |
| <partinfo>BBa_K300010</partinfo> | I80 | pSB1C3 | 25 |
| <partinfo>BBa_K300007</partinfo> | I70 - BioBrick | <partinfo>BBa_K300001</partinfo> | 26 |
| <partinfo>BBa_K300007</partinfo> | I70 - vector | <partinfo>BBa_K300001</partinfo> | 27 |
| <partinfo>BBa_K300004</partinfo> | INTEIN | pSB1C3 | 28 |
| <partinfo>BBa_I52002</partinfo> | I51 | <partinfo>BBa_K300000</partinfo> | 29 |
The parts we have in freezer and we only submitted to the Registry are:
| Part name | Wiki name | Plasmid |
|---|
| <partinfo>BBa_K300021</partinfo> | I8 | <partinfo>BBa_J61002</partinfo> |
| <partinfo>BBa_K300022</partinfo> | I9 | <partinfo>BBa_J61002</partinfo> |
| <partinfo>BBa_K300023</partinfo> | I10 | <partinfo>BBa_J61002</partinfo> |
| <partinfo>BBa_K300091</partinfo> | I63 | <partinfo>pSB1AK3</partinfo> |
| <partinfo>BBa_K300092</partinfo> | I64 | <partinfo>pSB1A2</partinfo> |
| <partinfo>BBa_K300099</partinfo> | I65 | <partinfo>pSB1A2</partinfo> |
| <partinfo>BBa_K300980</partinfo> | YEAST MR.G | pMA commercial vector |
| <partinfo>BBa_K300982</partinfo> | CREAM | - |
| <partinfo>BBa_K300983</partinfo> | CRIM MR.G | pMK-RQ commercial vector |
October, 19th
Wiki update and definition of results.
Every clones of E. coli genome integration project were amplified with PCR to verify their lengths and their positions into the genome.
In order to obtain better results we perform all the amplifications using an annealing temperature of 63°C.
Screening PCR was performed on MC42(A-B-C), MC43(A-B-C), MG42(B-C), MG43(A-B-C), MC1061 and MG1655. Primers used were BBa_K300975-BBa_K300976 (P1-P2), BBa_K300976-BBa_K300977 (P2-P3), BBa_K300977-BBa_K300978 (P3-P4).
 The order of the samples was the follow: MC42(A,B,C), MC43(A,B,C), MG42(B,C), MG43(A,B,C), blank, MC1061, MG1655. Primers were BBa_K300975-BBa_K300976 (P1-P2). 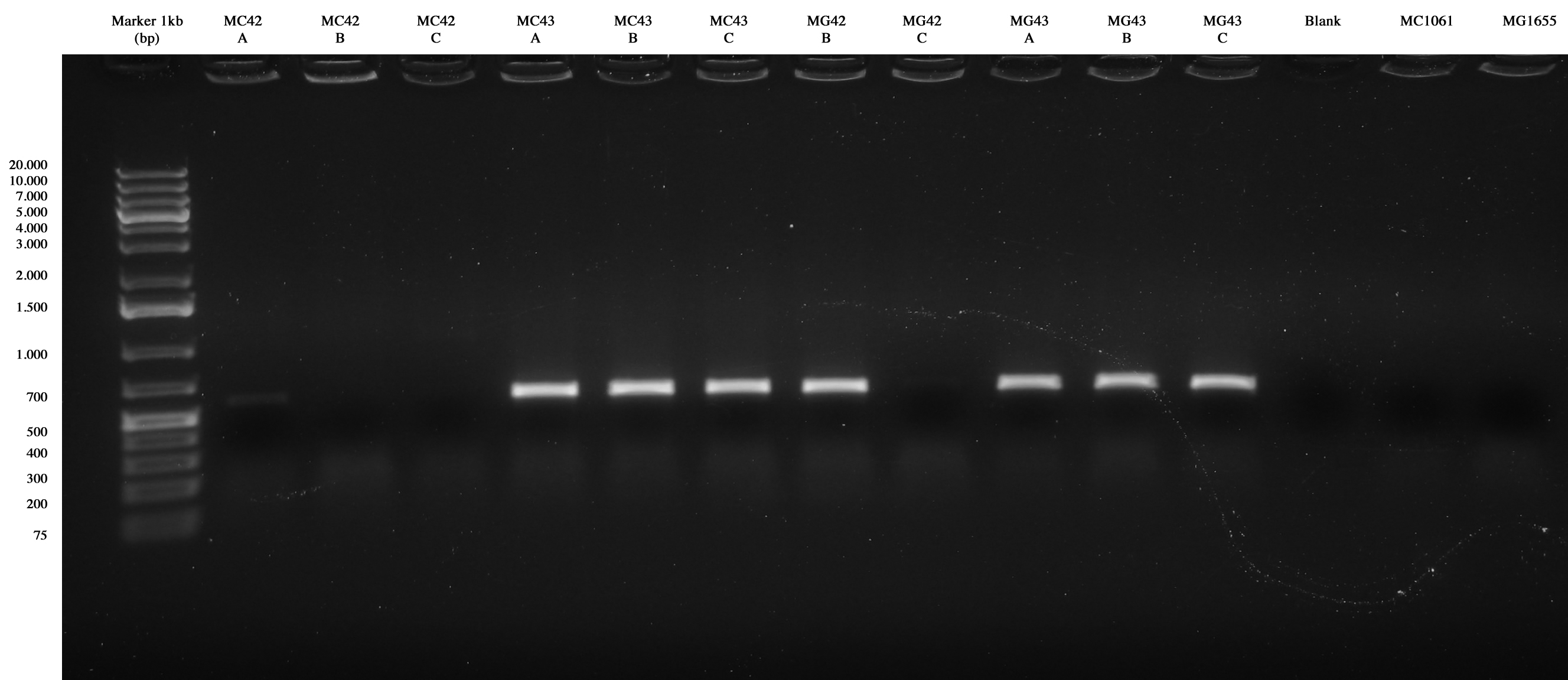 The order of the samples was the follow: MC42(A,B,C), MC43(A,B,C), MG42(B,C), MG43(A,B,C), blank, MC1061, MG1655. Primers were BBa_K300976-BBa_K300977 (P2-P3).  The order of the samples was the follow: MC42(A,B,C), MC43(A,B,C), MG42(B,C), MG43(A,B,C), blank, MC1061, MG1655. Primers were BBa_K300977-BBa_K300978 (P3-P4).
October, 20th
Wiki update and definition of results.
October, 21st
Wiki update and definition of results.
October, 22nd
Wiki update and definition of results.
October, 23rd
Wiki update and definition of results.
|
|
|
 "
"



