3A Assembly
From 2010.igem.org
(New page: __NOTOC__ ==Introduction== 3A assembly (which stands for three antibiotic assembly) is a method for assembling two BioBrick parts and selecting for correct assemblies with antibiotics.3A a...) |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| + | http://ginkgobioworks.com/support/ | ||
| + | |||
==Introduction== | ==Introduction== | ||
3A assembly (which stands for three antibiotic assembly) is a method for assembling two BioBrick parts and selecting for correct assemblies with antibiotics.3A assembly is now the preferred assembly method at iGEM HQ and the Knight lab at MIT. | 3A assembly (which stands for three antibiotic assembly) is a method for assembling two BioBrick parts and selecting for correct assemblies with antibiotics.3A assembly is now the preferred assembly method at iGEM HQ and the Knight lab at MIT. | ||
| - | It differs from | + | It differs from Standard Assembly in that it relies on three way ligation (between the two parts and the backbone vector) for assembly rather than two way ligation between a part and a part + vector. It uses effective antibiotic selection to eliminate unwanted background colonies and eliminates the need for gel purification and colony PCR of the resulting colonies. In theory, about 97% of the colonies should be the desired assembly. |
More recently, the Registry has been providing iGEM teams and Registry labs with linearized backbone plasmids to further improve this assembly method. This page has been updated to reflect the most modern approach. | More recently, the Registry has been providing iGEM teams and Registry labs with linearized backbone plasmids to further improve this assembly method. This page has been updated to reflect the most modern approach. | ||
| Line 46: | Line 48: | ||
==Procedure== | ==Procedure== | ||
#Miniprep your two parts. | #Miniprep your two parts. | ||
| - | #Digest your two parts and construction plasmid | + | #Digest your two parts and construction plasmid backbone destination vector with the following enzymes |
| - | #* | + | #*Upstream part with EcoRI and SpeI <br /> |
| - | + | {| border = "3" | |
| - | + | |- | |
| - | # | + | |align = "center" |Upstream Part Plasmid |
| - | #* | + | |align = "center" |500 ng |
| - | # | + | |- |
| - | #Transform the ligation product | + | |align = "center" |EcoRI-HF |
| + | |align = "center" |1 μL | ||
| + | |- | ||
| + | |align = "center" |SpeI | ||
| + | |align = "center" |1 μL | ||
| + | |- | ||
| + | |align = "center" |10X NEBuffer 2 | ||
| + | |align = "center" |5 μL | ||
| + | |- | ||
| + | |align = "center" |100X BSA | ||
| + | |align = "center" |0.5 μL | ||
| + | |- | ||
| + | |align = "center" |H2O | ||
| + | |align = "center" |to 50 μL | ||
| + | |- | ||
| + | |}<br /> | ||
| + | ** Downstream part with XbaI and PstI <br /> | ||
| + | {| border = "3" | ||
| + | |- | ||
| + | |align = "center" |Downstream Part Plasmid | ||
| + | |align = "center" |500 ng | ||
| + | |- | ||
| + | |align = "center" |XbaI | ||
| + | |align = "center" |1 μL | ||
| + | |- | ||
| + | |align = "center" |PstI | ||
| + | |align = "center" |1 μL | ||
| + | |- | ||
| + | |align = "center" |10X NEBuffer 2 | ||
| + | |align = "center" |5 μL | ||
| + | |- | ||
| + | |align = "center" |100X BSA | ||
| + | |align = "center" |0.5 μL | ||
| + | |- | ||
| + | |align = "center" |H2O | ||
| + | |align = "center" |to 50 μL | ||
| + | |- | ||
| + | |} <br /> | ||
| + | ** Destination plasmid backbone with EcoRI and PstI. Also digest the construction plasmid backbone with DpnI if possible to eliminate any plasmid remaining from the PCR. <br /> | ||
| + | {| border = "3" | ||
| + | |- | ||
| + | |align = "center" |Destination Plasmid Backbone | ||
| + | |align = "center" |500 ng | ||
| + | |- | ||
| + | |align = "center" |EcoRI-HF | ||
| + | |align = "center" |1 μL | ||
| + | |- | ||
| + | |align = "center" |SpeI | ||
| + | |align = "center" |1 μL | ||
| + | |- | ||
| + | |align = "center" |DpnI | ||
| + | |align = "center" |1 μL | ||
| + | |- | ||
| + | |align = "center" |10X NEBuffer 2 | ||
| + | |align = "center" |5 μL | ||
| + | |- | ||
| + | |align = "center" |100X BSA | ||
| + | |align = "center" |0.5 μL | ||
| + | |- | ||
| + | |align = "center" |H2O | ||
| + | |align = "center" |to 50 μL | ||
| + | |- | ||
| + | |} <br /> | ||
| + | #Purify the restriction digest. | ||
| + | #*For parts < 200 bp in length, do a [[Knight:Micropure EZ and Microcon purification]] (or alternatively an [[Ethanol precipitation of small DNA fragments]]). All other parts and the construction vector can be purified using a [http://www1.qiagen.com/HB/QIAquickSpin QIAquick PCR purification kit] or [http://www1.qiagen.com/literature/handbooks/PDF/DNACleanupAndConcentration/MinElute/1027886_HB_QQ_MinElute_0604.pdf| Qiagen Minelute PCR Purification Kit] (elutes in a smaller volume giving greater concentration of the DNA). | ||
| + | #Incubate all three restriction digest reactions at 37ºC for 10 minutes and then heat inactivate at 80ºC for 20 minutes. | ||
| + | #Ligation | ||
| + | {| border = "3" | ||
| + | |- | ||
| + | |align = "center" |Upstream Part digestion | ||
| + | |align = "center" |2 μL | ||
| + | |- | ||
| + | |align = "center" |Downstream Part digestion | ||
| + | |align = "center" |2 μL | ||
| + | |- | ||
| + | |align = "center" |Destination Backbone digestion | ||
| + | |align = "center" |2 μL | ||
| + | |- | ||
| + | |align = "center" |10X T4 DNA Ligase Buffer | ||
| + | |align = "center" |2 μL | ||
| + | |- | ||
| + | |align = "center" |T4 DNA Ligase | ||
| + | |align = "center" |0.5 μL | ||
| + | |- | ||
| + | |align = "center" |H2O | ||
| + | |align = "center" |11 μL | ||
| + | |- | ||
| + | |} | ||
| + | #Incubate at room temperature for 10 minutes and then heat inactivate at 80ºC for 20 minutes. | ||
| + | #Transform 2 μL of the ligation product into 50 μL of competent E. coli cells. Select using the antibiotic corresponding to the destination plasmid backbone. | ||
#If the input parts are good, almost all colonies will be correct. | #If the input parts are good, almost all colonies will be correct. | ||
#If desired analyze the transformation with single colony PCR followed by agarose gel electrophoresis. | #If desired analyze the transformation with single colony PCR followed by agarose gel electrophoresis. | ||
| Line 60: | Line 151: | ||
#Sequence the clone. | #Sequence the clone. | ||
# Record the sequence information in the Registry. | # Record the sequence information in the Registry. | ||
| + | |||
==Notes== | ==Notes== | ||
| Line 76: | Line 168: | ||
<br style="clear:both"/> | <br style="clear:both"/> | ||
*Since the transformed cells are plated on plates supplemented with the antibiotic corresponding to the construction plasmid, only cells containing the construction plasmid backbone should survive. | *Since the transformed cells are plated on plates supplemented with the antibiotic corresponding to the construction plasmid, only cells containing the construction plasmid backbone should survive. | ||
| - | *Since the construction plasmid itself has been reduced in quantity by PCR and digestion by | + | *Since the construction plasmid itself has been reduced in quantity by PCR and digestion by DpnI, few colonies will contain the original construction plasmid. |
==Contact== | ==Contact== | ||
This assembly technique was formulated by [[Randy Rettberg]] and implemented by Tom Knight and Reshma Shetty. | This assembly technique was formulated by [[Randy Rettberg]] and implemented by Tom Knight and Reshma Shetty. | ||
| + | |||
| + | Back to [[Team:Northwestern/Protocol|<span style="color:#4B088A;">Protocol</span>]] | ||
| + | |||
| + | [[Category:Protocol]] | ||
Latest revision as of 21:28, 16 October 2010
http://ginkgobioworks.com/support/
Introduction
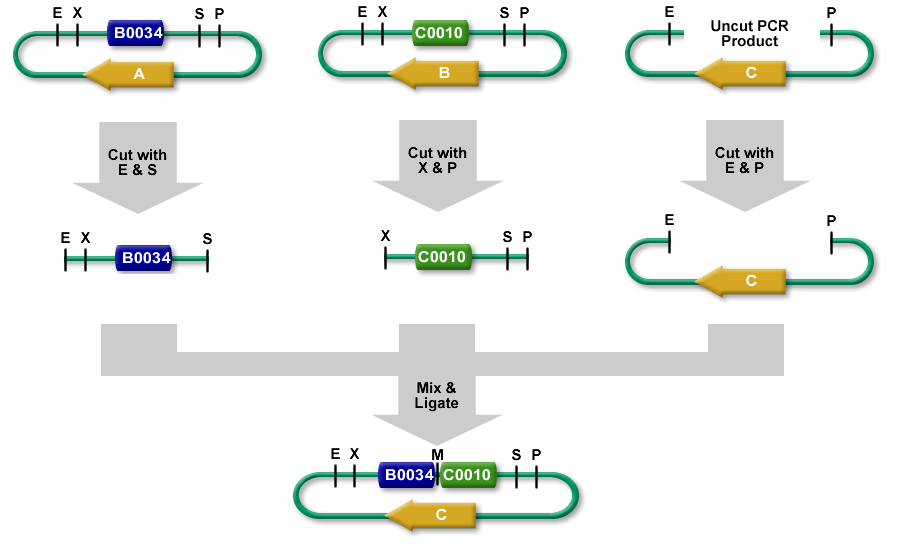
3A assembly (which stands for three antibiotic assembly) is a method for assembling two BioBrick parts and selecting for correct assemblies with antibiotics.3A assembly is now the preferred assembly method at iGEM HQ and the Knight lab at MIT.
It differs from Standard Assembly in that it relies on three way ligation (between the two parts and the backbone vector) for assembly rather than two way ligation between a part and a part + vector. It uses effective antibiotic selection to eliminate unwanted background colonies and eliminates the need for gel purification and colony PCR of the resulting colonies. In theory, about 97% of the colonies should be the desired assembly.
More recently, the Registry has been providing iGEM teams and Registry labs with linearized backbone plasmids to further improve this assembly method. This page has been updated to reflect the most modern approach.
Motivation
3A assembly was designed such that gel purification of the digested parts is unnecessary. Incorrect assemblies are selected against via either positive or negative selection. The motivations for eliminating the gel purification step are as follows
- Gel purification is slow and not easily susceptible to automation and scaling. As the Registry moves to larger scale assembly of parts, gel purification can be a limiting step in terms of how many assemblies can be done in parallel.
- Gel purification tends to be inefficient especially on small pieces of DNA. It can be difficult to isolate small pieces of DNA (< 200 bp) from a gel. Thus, when assembling two small pieces of DNA, 3A assembly might prove advantageous.
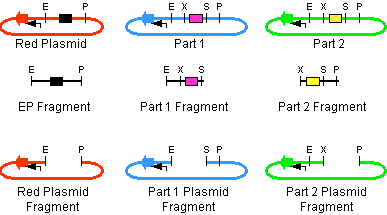
Description
- The left part is in a plasmid with an antibiotic resistance cassette for antibiotic A.
- The right part is in a plasmid with an antibiotic resistance cassette for antibiotic B.
- A construction plasmid is selected with neither resistance A nor B. It has antibiotic resistance C.
- The left part's plasmid is cut with EcoRI and SpeI.
- The right part's plasmid is cut with XbaI and PstI.
- The construction plasmid backbone is a linear piece of DNA. It has a few bases beyond the EcoRI and PstI restriction sites. It is cut with EcoRI and PstI.
- All 3 restriction digests are heated to heat kill all of the restriction enzymes.
- An equimolar quantity of all 3 restriction digest products are combined in a ligation reaction.
- The desired result is the left part 's SpeI overhang ligated with the right part's XbaI overhang resulting in a mixed site that cannot be cut with any of our enzymes.
- The new composite part is ligated into the construction plasmid backbone at the EcoRI and PstI sites.
- When the ligation result is transformed into cells and grown on plates with antibiotic C, only colonies with the correct construction(*) survive.
Materials
- Two parts in BioBricks format to be assembled. The parts are in a plasmid or they can be generated via PCR.
- A construction vector that has a different resistance marker from either of two parts.
- These construction backbones are available from the Registry.
- <partinfo>pSB1A3</partinfo>
- <partinfo>pSB1C3</partinfo>
- <partinfo>pSB1K3</partinfo>
- <partinfo>pSB1T3</partinfo>
Procedure
- Miniprep your two parts.
- Digest your two parts and construction plasmid backbone destination vector with the following enzymes
- Upstream part with EcoRI and SpeI
- Upstream part with EcoRI and SpeI
| Upstream Part Plasmid | 500 ng |
| EcoRI-HF | 1 μL |
| SpeI | 1 μL |
| 10X NEBuffer 2 | 5 μL |
| 100X BSA | 0.5 μL |
| H2O | to 50 μL |
- Downstream part with XbaI and PstI
- Downstream part with XbaI and PstI
| Downstream Part Plasmid | 500 ng |
| XbaI | 1 μL |
| PstI | 1 μL |
| 10X NEBuffer 2 | 5 μL |
| 100X BSA | 0.5 μL |
| H2O | to 50 μL |
- Destination plasmid backbone with EcoRI and PstI. Also digest the construction plasmid backbone with DpnI if possible to eliminate any plasmid remaining from the PCR.
- Destination plasmid backbone with EcoRI and PstI. Also digest the construction plasmid backbone with DpnI if possible to eliminate any plasmid remaining from the PCR.
| Destination Plasmid Backbone | 500 ng |
| EcoRI-HF | 1 μL |
| SpeI | 1 μL |
| DpnI | 1 μL |
| 10X NEBuffer 2 | 5 μL |
| 100X BSA | 0.5 μL |
| H2O | to 50 μL |
- Purify the restriction digest.
- For parts < 200 bp in length, do a Knight:Micropure EZ and Microcon purification (or alternatively an Ethanol precipitation of small DNA fragments). All other parts and the construction vector can be purified using a [http://www1.qiagen.com/HB/QIAquickSpin QIAquick PCR purification kit] or [http://www1.qiagen.com/literature/handbooks/PDF/DNACleanupAndConcentration/MinElute/1027886_HB_QQ_MinElute_0604.pdf| Qiagen Minelute PCR Purification Kit] (elutes in a smaller volume giving greater concentration of the DNA).
- Incubate all three restriction digest reactions at 37ºC for 10 minutes and then heat inactivate at 80ºC for 20 minutes.
- Ligation
| Upstream Part digestion | 2 μL |
| Downstream Part digestion | 2 μL |
| Destination Backbone digestion | 2 μL |
| 10X T4 DNA Ligase Buffer | 2 μL |
| T4 DNA Ligase | 0.5 μL |
| H2O | 11 μL |
- Incubate at room temperature for 10 minutes and then heat inactivate at 80ºC for 20 minutes.
- Transform 2 μL of the ligation product into 50 μL of competent E. coli cells. Select using the antibiotic corresponding to the destination plasmid backbone.
- If the input parts are good, almost all colonies will be correct.
- If desired analyze the transformation with single colony PCR followed by agarose gel electrophoresis.
- In rolling, large scale assembly, this step is often omitted.
- Miniprep clones that generated a band of the appropriate size.
- Sequence the clone.
- Record the sequence information in the Registry.
Notes
Obtaining materials
- All listed vectors are available to iGEM teams and Registry labs and courses through the [http://partsregistry.org Registry of Standard Biological Parts].
- As of spring 2010, we recommend using linear construction plasmid backbones that are available from the Registry or that you make yourself using PCR.
Troubleshooting
- In our experience, most problems with 3A Assembly come from the transformation process. Because 3A Assembly uses a triple ligation, the concentration of assembled DNA is about 10% of that produced by a double ligation process. Particularly, if your lab makes its own competent cells (and particularly chemically competent cells) it is important to test the competency. If you are having trouble obtaining colonies, we suggest that you try commercial competent cells. Be sure to follow the instructions carefully. Do not add more ligation product than specified and be careful when resuspending the competent cells.
- The second most common problems are caused by defects in the DNA entering the ligation. We find ligation to be a very reliable process. However, the sticky ends of the cut parts can be destroyed by enzymes. Also, the original DNA may be incorrect or defective.
- Another common mechanism for failure involves a piece of genomic DNA being cloned into the construction plasmid. This result typically occurs when the assembly is somehow unfavorable to the cell. This issue can be partially checked via the colony PCR step in the protocol though if the genomic DNA is of the same size as your assembly, then the single colony PCR screen is ineffective. A careful purification of the construction plasmid can help to avoid this result as can phosphatase treatment of the linear construction plasmid.
Analysis
- Since the transformed cells are plated on plates supplemented with the antibiotic corresponding to the construction plasmid, only cells containing the construction plasmid backbone should survive.
- Since the construction plasmid itself has been reduced in quantity by PCR and digestion by DpnI, few colonies will contain the original construction plasmid.
Contact
This assembly technique was formulated by Randy Rettberg and implemented by Tom Knight and Reshma Shetty.
Back to Protocol
 "
"