Team:Northwestern/Project
From 2010.igem.org
(Difference between revisions)
| (30 intermediate revisions not shown) | |||
| Line 101: | Line 101: | ||
<style> | <style> | ||
body { | body { | ||
| - | background: # | + | background: #EFCDF8; <!-- EEDCC8 url(https://static.igem.org/mediawiki/2010/3/38/BG1.jpg) repeat-y; --> |
} | } | ||
</style> | </style> | ||
| Line 110: | Line 110: | ||
<!--- The Mission, Experiments ---> | <!--- The Mission, Experiments ---> | ||
| - | {|- style="font-size: 87%; background: # | + | {|- style="font-size: 87%; background: #EFCDF8; color: #FFFFFF" cellpadding="1" cellspacing="1" bordercolor="#000000" width="75%" align="center" |
| - | !align="center"|[[Team:Northwestern|<font color="# | + | !align="center"|[[Team:Northwestern|<font color="#2B3856">'''Home'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Brainstorm|<font color="# | + | !align="center"|[[Team:Northwestern/Brainstorm|<font color="#000000">'''Brainstorm'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Team|<font color="# | + | !align="center"|[[Team:Northwestern/Team|<font color="#000000">'''Team'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Acknowledgements|<font color="# | + | !align="center"|[[Team:Northwestern/Acknowledgements|<font color="#000000">'''Acknowledgements'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Project|<font color="# | + | !align="center"|[[Team:Northwestern/Project|<font color="#000000">'''Project'''</font>]] |
| - | !align="center"|[[Team:Northwestern/SideProject|<font color="# | + | !align="center"|[[Team:Northwestern/SideProject|<font color="#000000">'''Human Practices'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Parts|<font color="# | + | !align="center"|[[Team:Northwestern/Parts|<font color="#000000">'''Parts'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Notebook|<font color="# | + | !align="center"|[[Team:Northwestern/Notebook|<font color="#000000">'''Notebook'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Calendar|<font color="# | + | !align="center"|[[Team:Northwestern/Calendar|<font color="#000000">'''Calendar'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Protocol|<font color="# | + | !align="center"|[[Team:Northwestern/Protocol|<font color="#000000">'''Protocol'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Safety|<font color="# | + | !align="center"|[[Team:Northwestern/Safety|<font color="#000000">'''Safety'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Links|<font color="# | + | !align="center"|[[Team:Northwestern/Links|<font color="#000000">'''Links'''</font>]] |
| - | !align="center"|[[Team:Northwestern/References|<font color="# | + | !align="center"|[[Team:Northwestern/References|<font color="#000000">'''References'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Media|<font color="# | + | !align="center"|[[Team:Northwestern/Media|<font color="#000000">'''Media'''</font>]] |
| - | !align="center"|[[Team:Northwestern/Contact|<font color="# | + | !align="center"|[[Team:Northwestern/Contact|<font color="#000000">'''Contact'''</font>]] |
|} | |} | ||
| - | {| align="center" border="0" width="75%" | + | {| align="center" border="0" width="75%" cellpadding="20" |
| - | + | |colspan="2" style="padding:0"| | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<html> | <html> | ||
<table align="center"> | <table align="center"> | ||
<tr> | <tr> | ||
| + | <td><a href="https://2010.igem.org/Team:Northwestern/Project/Modeling"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/a/a7/NU_modeling.jpg"></a></td> | ||
<td><a href="https://2010.igem.org/Team:Northwestern/Project/Chassis"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/a/a6/Nuchassis.jpg"></a></td> | <td><a href="https://2010.igem.org/Team:Northwestern/Project/Chassis"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/a/a6/Nuchassis.jpg"></a></td> | ||
<td><a href="https://2010.igem.org/Team:Northwestern/Project/Lac"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/1/18/Nulac.jpg"></a></td> | <td><a href="https://2010.igem.org/Team:Northwestern/Project/Lac"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/1/18/Nulac.jpg"></a></td> | ||
<td><a href="https://2010.igem.org/Team:Northwestern/Project/Chitin Synthesis"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/4/4e/Nuchitin.jpg"></a></td> | <td><a href="https://2010.igem.org/Team:Northwestern/Project/Chitin Synthesis"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/4/4e/Nuchitin.jpg"></a></td> | ||
<td><a href="https://2010.igem.org/Team:Northwestern/Project/Apoptosis"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/c/cb/Nuapop.jpg"></a></td> | <td><a href="https://2010.igem.org/Team:Northwestern/Project/Apoptosis"><img width="100" class="icon" src="https://static.igem.org/mediawiki/2010/c/cb/Nuapop.jpg"></a></td> | ||
| - | |||
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <td align="center">Modeling</td> | ||
<td align="center">Chassis</td> | <td align="center">Chassis</td> | ||
<td align="center">Induction</td> | <td align="center">Induction</td> | ||
<td align="center">Chitin</td> | <td align="center">Chitin</td> | ||
<td align="center">Apoptosis</td> | <td align="center">Apoptosis</td> | ||
| - | |||
</tr> | </tr> | ||
</table> | </table> | ||
</html> | </html> | ||
| - | |||
|- | |- | ||
| - | | | + | |align="left" valign="top" width="80%" cellpadding="10"| |
| + | =='''Overview'''== | ||
| + | Our main project is to simulate mammalian skin using chitin in E. Coli. | ||
| + | |||
| + | In mammalian skin, mitosis occurs in the basal layer of the epithelial cells and cells travel outwards towards the surface of the skin as they mature. As the cells move further away from the basal layer, they die and their cytoplasm is released and the cells are filled with keratin, thus forming a continuously regenerating protective layer on the outer-most part of epithelial layer; our project is modeled after this. | ||
| + | |||
| + | |||
| + | First, a specific strain of Escherichia Coli is used to generate a thick bacterial biofilm lawn, which is formed on an enriched LB agar plate. The strain also does not produce chitinase so that the produced chitin is not digested. | ||
| + | |||
| + | |||
| + | Then a fast, high-expression signalling pathway initiates production of chitin in the top-most layer of the lawn. The gene for chitin synthesis is extracted from the genome of Saccharomyces Cerevisiae, more commonly known as Baker's Yeast, while the signalling pathway and expression parts are identified from the parts registry and extracted from the iGEM 2010 Distribution kit. | ||
| + | |||
| + | |||
| + | Simultaneously, a slow, medium-expression signalling pathway initiates production of a cell lysis expression vector in the top-most layer of the lawn. The apoptosis part used is the T4 Bacteriophage apoptosis cassette, and this part as well as the signalling and expression parts, are identified from the parts registry and extracted from the iGEM 2010 Distribution kit. | ||
| + | |||
| + | |||
| + | With both vectors, the E.Coli cells in the top layer generates chitin then lyses, thus depositing a layer of chitin in the top-most layer of the bacterial lawn. | ||
| + | |||
| + | |||
| + | To identify the expression of chitin, a chitin-specific rhodamine stain is used. To identify the efficacy of the apoptosis cassette, a live-dead assay is used. With these stains, the colony can be imaged vertically through confocal microscopy to show the deposition of Chitin in the top layer. | ||
| + | |||
| + | |||
| + | Whenever there is damage to the chitin layer, the signalling pathway can be reinitiated to regenerate chitin in the damaged location. | ||
| + | |||
| + | |||
| + | ||[[Image:PosterChild.jpg|400px|center]] | ||
| + | |- | ||
| + | |colspan="2" cellpadding="10"| | ||
==Strategy== | ==Strategy== | ||
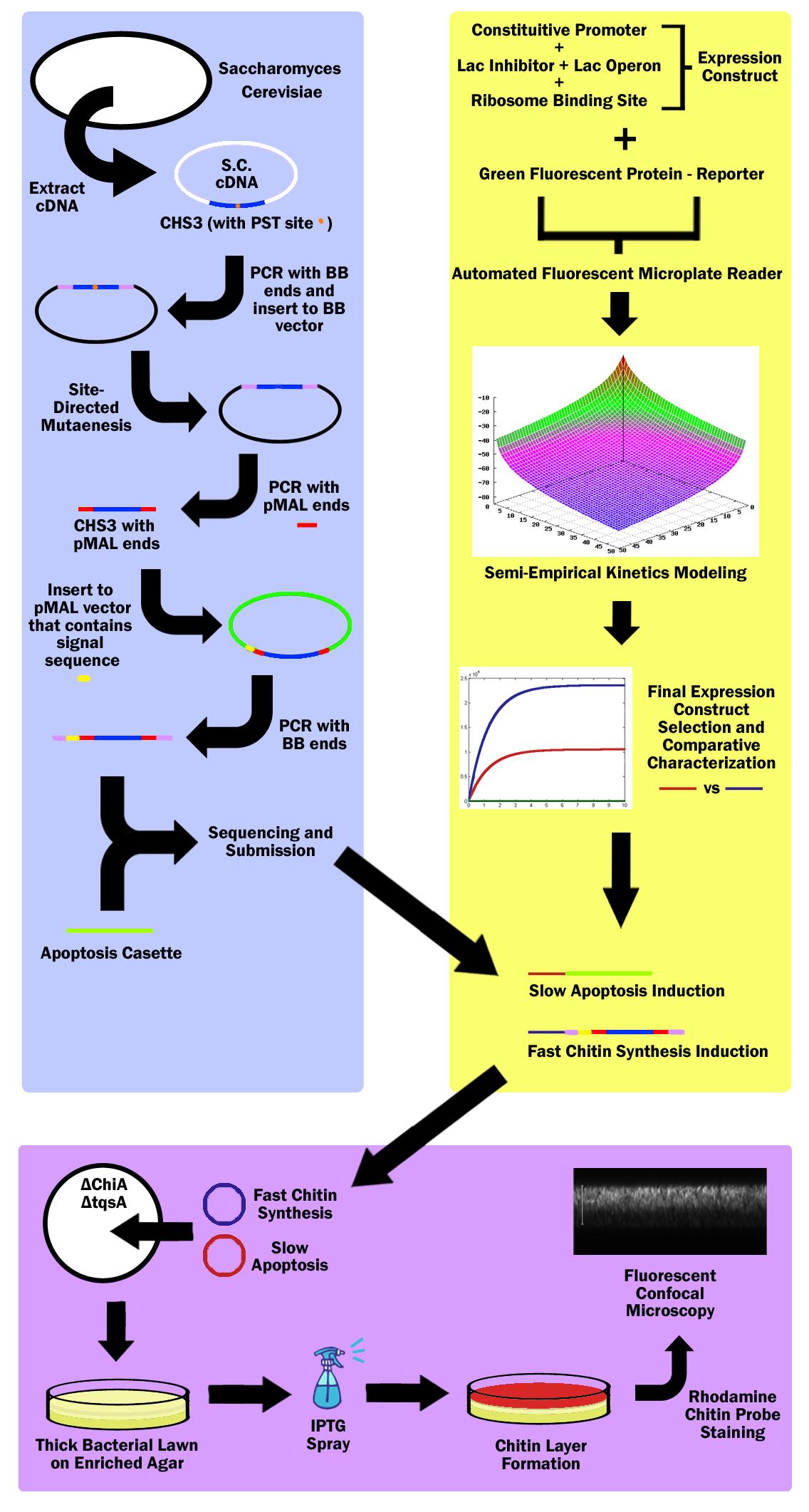
===Obtaining Chitin Synthase 3 Gene=== | ===Obtaining Chitin Synthase 3 Gene=== | ||
| - | First, the Chitin Synthase 3 (CHS3) gene | + | First, the Chitin Synthase 3 (CHS3) gene is subcloned out of Saccharomyces Cerevisiae (Baker’s Yeast) cDNA (complementary DNA – contains no introns, thus can be used in prokaryotes) with biobrick restriction sites (EcoR1, Xbal1 on one end, and Spe1, Pst1 on the other). CHS3 was chosen as it was found to be the major enzyme (knockouts had 80% reduced Chitin) in its family and requires no co-enzyme or activating compounds. Upon running CHS3 through NEB Cutter V2.0, we found a PST1 restriction site which is incompatible with the biobrick standard (EcoR1, Xbal, Spe1, Pst1). To remove this Pst1 site, we insert CHS3 into a biobrick vector plasmid, and performed site-directed mutagenesis (SDM). |
===pMAL Vector=== | ===pMAL Vector=== | ||
| - | Research revealed that chitin synthase is a transmembrane enzyme, and in order to simulate similar conditions, we decided to use the pMAL vector by NEB which contains a periplasmic membrane signal sequence. CHS3 | + | Research revealed that chitin synthase is a transmembrane enzyme, and in order to simulate similar conditions, we decided to use the pMAL vector by NEB which contains a periplasmic membrane signal sequence. CHS3 is subcloned in to the pMAL-p5x NEB vector by first PCR’ing it out of the biobrick vector plasmid - on which it must undergo SDM - with pMAL restriction sites (Nde1 and EcoR1) and ligated into the pMAL-p5x vector. Then, the CHS3 is again amplified with biobrick sites, but this time, with the periplasmic signal sequence native to the pMAL-p5x plasmid, and inserted into a standard biobrick vector. The entire part is then sequenced and submitted to the registry of standard parts. |
| - | A bacterial apoptosis cassette made by the Brown iGEM team in ‘08 | + | A bacterial apoptosis cassette made by the Brown iGEM team in ‘08 is taken from the iGEM 2010 distribution kit and colony amplified. |
===Constitutive Promoters & Lac Operon=== | ===Constitutive Promoters & Lac Operon=== | ||
| - | 3 constitutive promoters and 2 lac-inhibitor/lac-operon combination parts | + | 3 constitutive promoters and 2 lac-inhibitor/lac-operon combination parts are taken from the registry and ligated in all 6 combinations through 3A Assembly. These combinations express different levels of lacI, thus exhibiting variable inductivity. These 6 constructs are then ligated with a reporter protein - green fluorescent protein - and characterized via an automated fluorescent microplate reader; fluorescence is used to determine the construct inductivity and expression level. |
===IPTG Activation=== | ===IPTG Activation=== | ||
| - | + | The data is used to fit our own theoretical kinetics model that accounts for IPTG diffusion through biofilm, external IPTG concentration, internal IPTG concentration, lac repressor concentration, repressor bound gene (DNA) concentration, unbound gene (DNA) concentration, mRNA concentration, enzyme concentration, substrate concentration, enzyme-substrate complex concentration, and final product concentration, as well as all the rate constants involved. Using the data acquired from the microplate reader, a final semi-empirical kinetics model is generated. Using this model, two different constructs are chosen; both with similar inductivity, but one with and the other without time delay. | |
===Chitin Synthesis Production=== | ===Chitin Synthesis Production=== | ||
| - | The fast expression construct | + | The fast expression construct is ligated with Chitin Synthase, while the slow expression construct is ligated with the apoptosis cassette. Both plasmids are then transformed into a chiA and tqsA double knockout strain of K12 Escherichia Coli. ChiA knockouts lack Chitinase, which is an enzyme that catabolizes chitin. tqsA knockouts have a disrupted quorum sensing mechanism that allows them to grow very thick lawns. The mutants are acquired from Keio collection (one from Northwestern, and the other from Yale respectively). |
| + | |||
| + | ===MAGE=== | ||
| + | The Multiplex Automated Genome Engineering (MAGE) protocol developed by H. H. Wang. et al. is used to acquire double knockouts. The MAGE protocol is an accelerated evolution procedure where oligonucleotides are electroporated into the cells in order to introduce mutations in the genome during replication by binding as lagging strand primers, taking advantage of the lambda-red construct. | ||
| + | |||
===Mechanism=== | ===Mechanism=== | ||
| - | The double knockout colonies with both constructs | + | The double knockout colonies with both constructs are then sprayed with IPTG, which first induces the fast chitin synthesis construct, causi a buildup of chitin in cells in the top layer of the bacterial lawn. Then the slow apoptosis construct is induced, which then lyses the cells, and releases a layer of chitin in the top layer of the bacterial lawn. This construct is then stained with a chitin-specific rhodamine probe by NEB as well as Baclight by Invitrogen in order to ensure that the cells are producing chitin and lysing. The lawn is then imaged through a fluorescent confocal microscope. |
|} | |} | ||
Latest revision as of 23:58, 23 October 2010
| Home | Brainstorm | Team | Acknowledgements | Project | Human Practices | Parts | Notebook | Calendar | Protocol | Safety | Links | References | Media | Contact |
|---|
 "
"