Team:SDU-Denmark/labnotes8
From 2010.igem.org
(→Lab notes (8/30 - 9/5)) |
(→Lab notes (8/30 - 9/5)) |
||
| (6 intermediate revisions not shown) | |||
| Line 270: | Line 270: | ||
<br> | <br> | ||
Results: Primers were confirmed working, the next step will be PFU pcr.<br> | Results: Primers were confirmed working, the next step will be PFU pcr.<br> | ||
| - | |||
== Retinal == | == Retinal == | ||
| - | === | + | === Gradient PCR on ninaB (after PCR with ninaB2fw and ninaB2rv) with ninaBfw and ninaBrv === |
| - | Date: 30/ | + | Date: 30/08<br> |
| - | Done by: Tommy<br> | + | Done by: Tommy & Marie<br> |
Methods: PCR<br> | Methods: PCR<br> | ||
| - | Protocols: CP1. | + | Protocols: CP1.1[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1]<br> |
Notes: <br> | Notes: <br> | ||
| - | + | To finde the optimal anneling temperatur a gradient PCR from 60 to 75 degrees<br> | |
| - | + | "ninaB" was used as template and ninaB2fw and ninaB2rv was used as primers<br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
PCR Program:<br> | PCR Program:<br> | ||
| Line 309: | Line 304: | ||
<td style="vertical-align: top;">Annealing<br> | <td style="vertical-align: top;">Annealing<br> | ||
</td> | </td> | ||
| - | <td style="vertical-align: top;"> | + | <td style="vertical-align: top;">Grad. C<br> |
</td> | </td> | ||
<td style="vertical-align: top;">1 min<br> | <td style="vertical-align: top;">1 min<br> | ||
| Line 319: | Line 314: | ||
<td style="vertical-align: top;">72 C<br> | <td style="vertical-align: top;">72 C<br> | ||
</td> | </td> | ||
| - | <td style="vertical-align: top;"> | + | <td style="vertical-align: top;">4 min<br> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 347: | Line 342: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | + | Vary small amount of the product are observed, it is pooled and gel purifid, the nanodrop shows 372,3 and 377,1 ng/uL. | |
| - | === | + | === Gel purification af ninaB from gradient PCR === |
| - | Date: | + | Date: 31/08<br> |
Done by: Tommy & Marie<br> | Done by: Tommy & Marie<br> | ||
| - | Methods: | + | Methods: PCR<br> |
| - | Protocols: | + | Protocols: CP1.1[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1]<br> |
Notes: <br> | Notes: <br> | ||
| - | + | Gel purification was preformed on the pooled product from the gradient PCR according to the protocol<br> | |
| - | + | After the gel purification the 2 samples was pooled and nanodroped: 7.0 ng/ul<br> | |
| - | + | Furthere PCR was preformed on the purifid product<br> | |
| - | === | + | |
| - | Date: | + | === Gel purification af ninaB from gradient PCR === |
| - | Done by: Tommy<br> | + | Date: 1/09<br> |
| - | Methods: | + | Done by: Tommy & Marie<br> |
| + | Methods: PCR<br> | ||
Protocols: CP1.3[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.3]<br> | Protocols: CP1.3[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.3]<br> | ||
Notes: <br> | Notes: <br> | ||
| - | + | PCR was run according to protocol to the following to this program<br> | |
| - | + | Gel purified produckt was used as template and ninaBfw and ninaBrv was used as primers<br> | |
| - | + | ||
| - | + | ||
| - | + | ||
PCR Program:<br> | PCR Program:<br> | ||
<table style="text-align: left;" border="1" cellpadding="2" | <table style="text-align: left;" border="1" cellpadding="2" | ||
| Line 392: | Line 385: | ||
<td style="vertical-align: top;">Annealing<br> | <td style="vertical-align: top;">Annealing<br> | ||
</td> | </td> | ||
| - | <td style="vertical-align: top;"> | + | <td style="vertical-align: top;">Grad. C<br> |
</td> | </td> | ||
<td style="vertical-align: top;">1 min<br> | <td style="vertical-align: top;">1 min<br> | ||
| Line 402: | Line 395: | ||
<td style="vertical-align: top;">72 C<br> | <td style="vertical-align: top;">72 C<br> | ||
</td> | </td> | ||
| - | <td style="vertical-align: top;"> | + | <td style="vertical-align: top;">4 min<br> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 410: | Line 403: | ||
<td style="vertical-align: top;">rep<br> | <td style="vertical-align: top;">rep<br> | ||
</td> | </td> | ||
| - | <td style="vertical-align: top;"> | + | <td style="vertical-align: top;">44x<br> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 430: | Line 423: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | === | + | === Restriction digest of ninaB === |
| - | Date: 2/ | + | Date: 2/09<br> |
| - | Done by: Tommy<br> | + | Done by: Tommy & Marie<br> |
| - | Methods: | + | Methods: restriction digest<br> |
| - | Protocols: | + | Protocols: RD1.1[https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1]<br> |
Notes: <br> | Notes: <br> | ||
| - | + | EcoRI and PstI ristriction enzymes was used and the ristriction digest was preformed according to protocol<br> | |
| - | + | No gel purification was preformed, purificatin was preformed directly from the restriction mixture<br> | |
| - | + | Purification was deno using the GFX purification kit and proformed according to that protocol, the purifid product was nanodroped: 3,1 ng/uL | |
| - | + | ||
| - | === | + | === "Cross-PCR" on ninaB === |
| - | Date: 2/ | + | Date: 2/09<br> |
| - | Done by: Tommy<br> | + | Done by: Tommy & Marie<br> |
| - | Methods: | + | Methods: PCR<br> |
| - | Protocols: | + | Protocols: CP1.1[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1]<br> |
Notes: <br> | Notes: <br> | ||
| - | + | Tubes A: ninaBfw and ninaB2rv<br> | |
| - | + | Tubes B: ninaB2fw and ninaBrv<br> | |
| - | + | NinaB PCR product with ninaB2fw and rv was used as template, both the tubes was run on the same PCR program | |
| + | <table style="text-align: left;" border="1" cellpadding="2" | ||
| + | cellspacing="2"> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Start<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">94 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">2 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Denaturing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">94 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Annealing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">68 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Elongation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">4 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Goto2<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">rep<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">44x<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">End<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">5 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Hold<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">4 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;"><br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | === Miniprep of J13002 === | |
| - | + | Date: 3/09<br> | |
| - | + | Done by: Tommy & Marie<br> | |
| + | Methods: Miniprep<br> | ||
| + | Protocols: MP1.1[https://2010.igem.org/Team:SDU-Denmark/protocols#MP1.1]<br> | ||
| + | Notes: <br> | ||
| + | The minipreps was done according to protocol<br> | ||
| + | Nanodrop:<br> | ||
| + | Tube 1: 38,9 ng/ul, 2nd elution: 20,5 ng/ul <br> | ||
| + | Tube 2: 36,9 ng/ul, 2nd elution: 18,7 ng/ul <br> | ||
| + | Tube 3: 38,4 ng/ul, 2nd elution: 20,0 ng/ul <br> | ||
| + | Tube 4: 32,6 ng/ul, 2nd elution: 16,4 ng/ul <br> | ||
| - | + | The pooled samples was nanodroped: 20,6 ng/uL | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 19:21, 16 October 2010
Lab notes (8/30 - 9/5)
Contents |
Flagella
Since the previous FlhDCmut was wrongly mutated due to incorrect mutation primers, we are now back to square one with new correct primers. The next weeks the flagella-group are working according to the following plan:
1) Miniprep of plasmids with "wrong" FlhDCmut
2) Two-step PCR to get mutatet FlhDC
3) Cut and Ligate into pSB1C3 and pSB1AK3 and transform into TOP10 cells
4) Send to sequencing
5) Characterize biobrick
Miniprep of "wrong" FlhDCmut
Done by: Louise
Date: September 3rd
Protocol: [MP1.2]
Notes:
No changes of protocol were made.
Results:
Nanodrop after sample was dried down:
Sample 1: Concentration: 192ng/ul Purity: 1.96/1.90
Sample 2: Concentration: 200ng/ul Purity: 1.84/1.88
The samples were run on a 1.5% gel with a 10kb ladder. The bands are positioned between 1500bp and 1200bp. FlhDCmut in pSB1C3 is 1248bp.
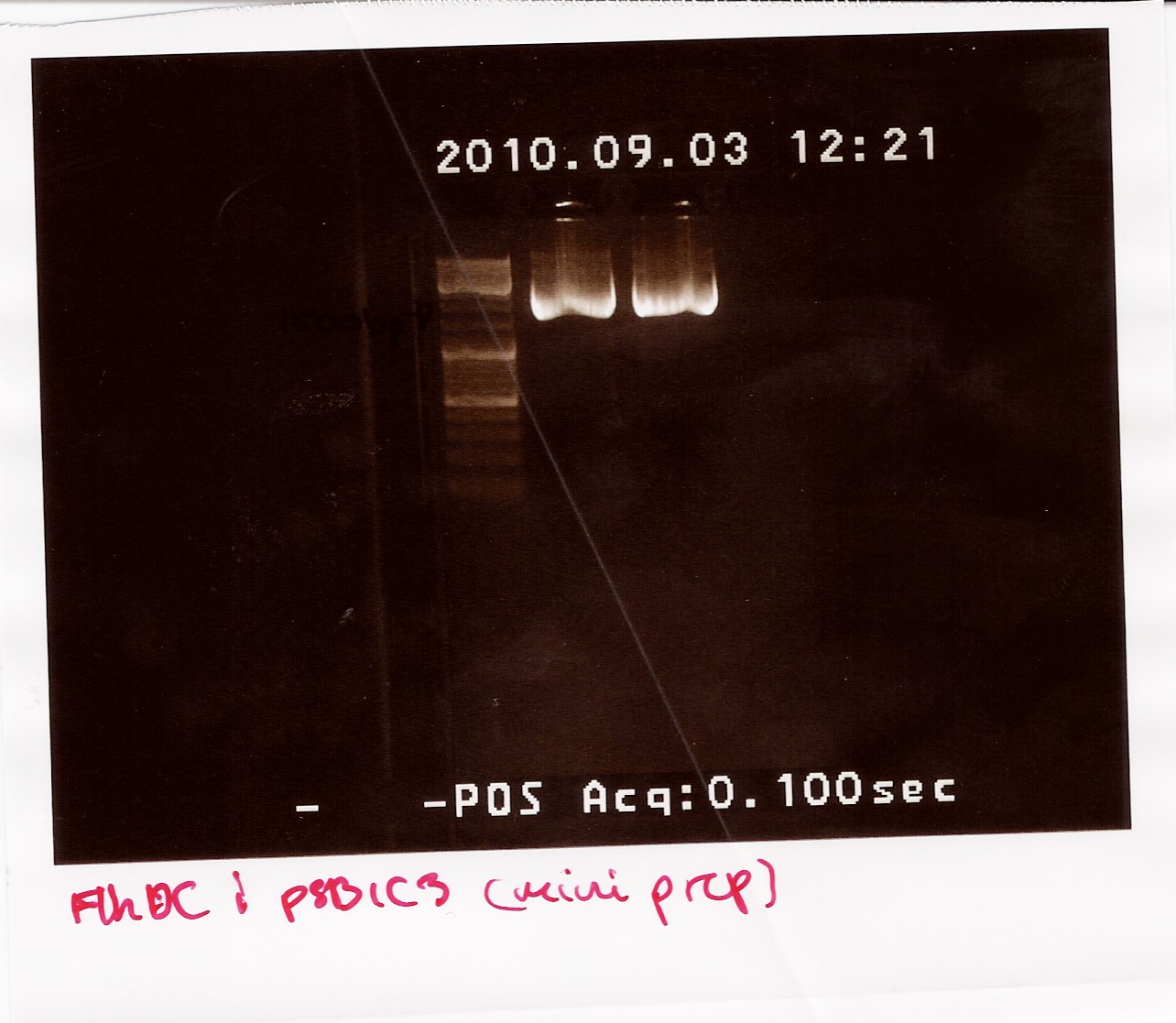
Photosensor
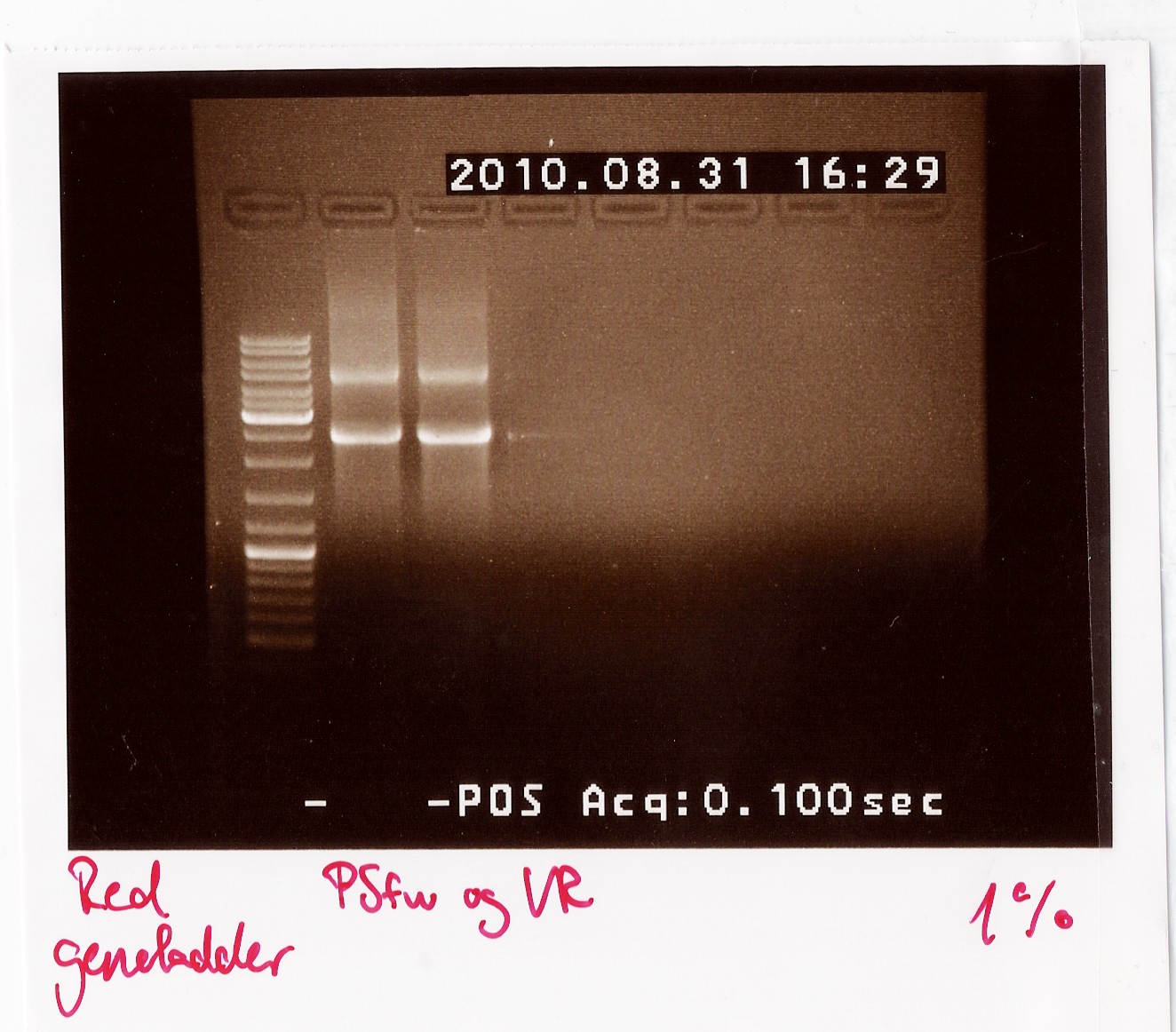
PCR on pKJ606 with PSfw and VR primers
Date: 31/8
https://2010.igem.org/wiki/index.php?title=Team:SDU-Denmark/labnotes8&action=edit§ion=3
Done by: LC
Methods: PCR
Protocols: CP1.3[1]
Notes:
Premix:
7,5 µl 10xTAQ Buffer
3 µl MgCl2
3 µl VF2
3 µl VR
1,5 µl dNTP
55,5 µl H2O
3 µl template (PS1 miniprep)
3/8 µl TAQ Polymerase
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
3 min |
| Goto2 |
rep |
29x |
| End |
72 C |
3 min |
| Hold |
4 C |
Results: The bands that showed up were around 2500 BP as expected from the sequencing results. This means that the designed primers have a high possibility of working, so that they will be ordered.
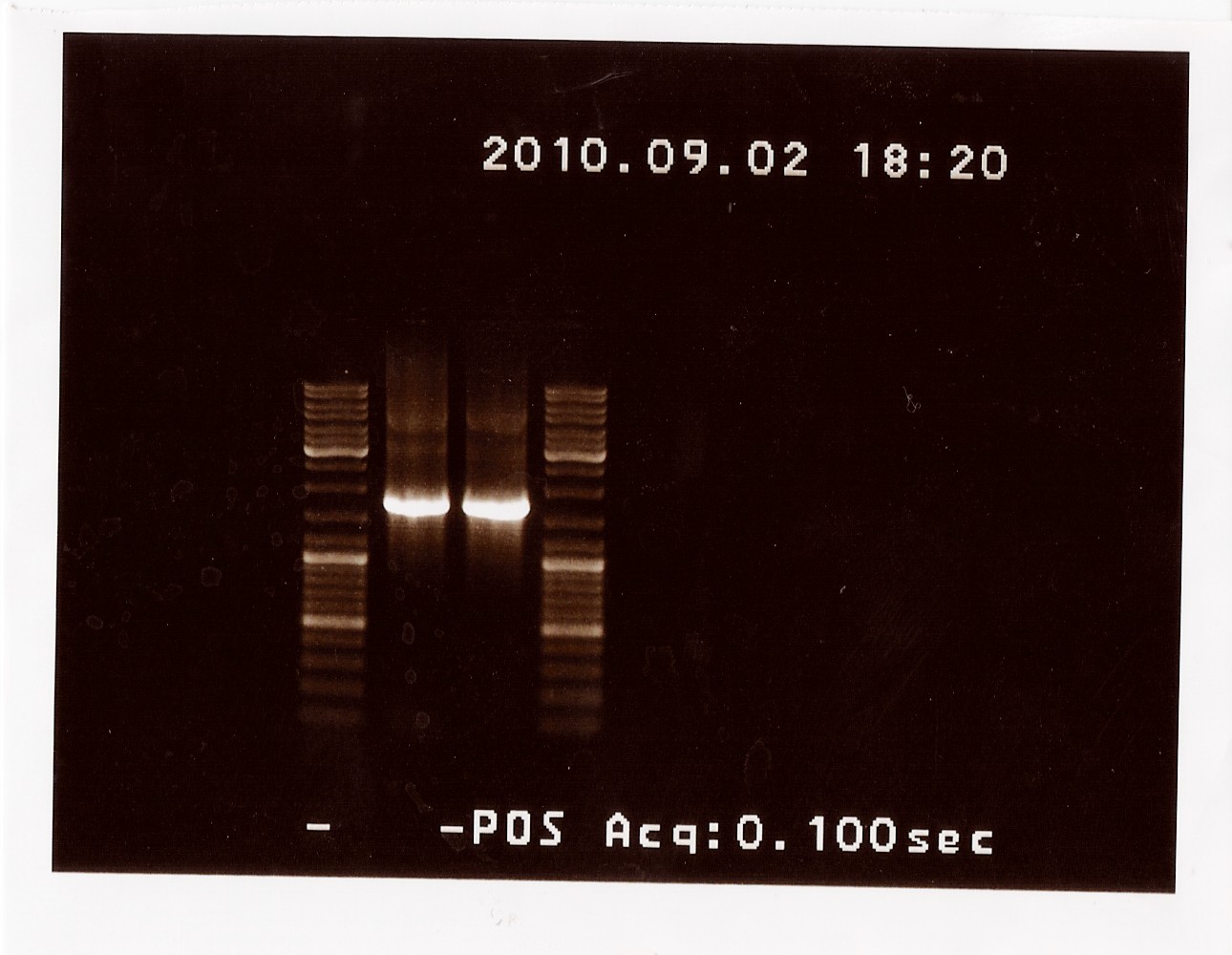
PCR on pKJ606 with PSfw and PSrv primers
Date: 02/09
Done by: LC
Methods: PCR
Protocols: CP1.3[2]
Notes:
Premix:
7,5 µl 10xTAQ Buffer
3 µl MgCl2
3 µl VF2
3 µl VR
1,5 µl dNTP
55,5 µl H2O
3 µl template (PS1 miniprep)
3/8 µl TAQ Polymerase
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2 min |
| Goto2 |
rep |
29x |
| End |
72 C |
3 min |
| Hold |
4 C |
Results: The bands that showed up were around 1750 BP, further confirming sequencing results. New primers for taking the whole gene out have been ordered.
PCR on pKJ606 with fwPS2 and rvPS2 primers
Date: 04/09
Done by: LC
Methods: PCR
Protocols: CP1.3[3]
Notes:
Premix:
12,5 µl 10xTAQ Buffer
5 µl MgCl2
5 µl VF2
5 µl VR
2,5 µl dNTP
59,5 µl H2O
4 µl template (Consisting of 2 µl H2O and 2 µl of pKJ606 miniprep)
1/2 µl TAQ Polymerase
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2:30 min |
| Goto2 |
rep |
29x |
| End |
72 C |
3 min |
| Hold |
4 C |
Results: Primers were confirmed working, the next step will be PFU pcr.
Retinal
Gradient PCR on ninaB (after PCR with ninaB2fw and ninaB2rv) with ninaBfw and ninaBrv
Date: 30/08
Done by: Tommy & Marie
Methods: PCR
Protocols: CP1.1[4]
Notes:
To finde the optimal anneling temperatur a gradient PCR from 60 to 75 degrees
"ninaB" was used as template and ninaB2fw and ninaB2rv was used as primers
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
Grad. C |
1 min |
| Elongation |
72 C |
4 min |
| Goto2 |
rep |
29x |
| End |
72 C |
5 min |
| Hold |
4 C |
Vary small amount of the product are observed, it is pooled and gel purifid, the nanodrop shows 372,3 and 377,1 ng/uL.
Gel purification af ninaB from gradient PCR
Date: 31/08
Done by: Tommy & Marie
Methods: PCR
Protocols: CP1.1[5]
Notes:
Gel purification was preformed on the pooled product from the gradient PCR according to the protocol
After the gel purification the 2 samples was pooled and nanodroped: 7.0 ng/ul
Furthere PCR was preformed on the purifid product
Gel purification af ninaB from gradient PCR
Date: 1/09
Done by: Tommy & Marie
Methods: PCR
Protocols: CP1.3[6]
Notes:
PCR was run according to protocol to the following to this program
Gel purified produckt was used as template and ninaBfw and ninaBrv was used as primers
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
Grad. C |
1 min |
| Elongation |
72 C |
4 min |
| Goto2 |
rep |
44x |
| End |
72 C |
5 min |
| Hold |
4 C |
Restriction digest of ninaB
Date: 2/09
Done by: Tommy & Marie
Methods: restriction digest
Protocols: RD1.1[7]
Notes:
EcoRI and PstI ristriction enzymes was used and the ristriction digest was preformed according to protocol
No gel purification was preformed, purificatin was preformed directly from the restriction mixture
Purification was deno using the GFX purification kit and proformed according to that protocol, the purifid product was nanodroped: 3,1 ng/uL
"Cross-PCR" on ninaB
Date: 2/09
Done by: Tommy & Marie
Methods: PCR
Protocols: CP1.1[8]
Notes:
Tubes A: ninaBfw and ninaB2rv
Tubes B: ninaB2fw and ninaBrv
NinaB PCR product with ninaB2fw and rv was used as template, both the tubes was run on the same PCR program
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
68 C |
1 min |
| Elongation |
72 C |
4 min |
| Goto2 |
rep |
44x |
| End |
72 C |
5 min |
| Hold |
4 C |
Miniprep of J13002
Date: 3/09
Done by: Tommy & Marie
Methods: Miniprep
Protocols: MP1.1[9]
Notes:
The minipreps was done according to protocol
Nanodrop:
Tube 1: 38,9 ng/ul, 2nd elution: 20,5 ng/ul
Tube 2: 36,9 ng/ul, 2nd elution: 18,7 ng/ul
Tube 3: 38,4 ng/ul, 2nd elution: 20,0 ng/ul
Tube 4: 32,6 ng/ul, 2nd elution: 16,4 ng/ul
The pooled samples was nanodroped: 20,6 ng/uL
 "
"