Team:KAIST-Korea/Project/Methods
From 2010.igem.org
(Difference between revisions)
| (16 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<tr> | <tr> | ||
<td valign="top" width="75%"> | <td valign="top" width="75%"> | ||
| - | <center><a href="2010.igem.org/Team:KAIST-Korea/Methods/PCR"><img src="https://static.igem.org/mediawiki/2010/ | + | <html> |
| - | + | <center> | |
| - | + | <a href="https://2010.igem.org/Team:KAIST-Korea/Project/Methods"><img src="https://static.igem.org/mediawiki/2010/6/6f/Method_button.png" width = 300></a> | |
| + | <a href="https://2010.igem.org/Team:KAIST-Korea/Project/Methods/PCR"><img src="https://static.igem.org/mediawiki/2010/e/e2/PCR-KAIST3.png" width = 300></a> | ||
| + | <a href="https://2010.igem.org/Team:KAIST-Korea/Project/Methods/PCR_Result"><img src="https://static.igem.org/mediawiki/2010/9/94/PCR-Exp.png" width = 300></a></center> | ||
| + | </html> | ||
| + | <br> | ||
| - | == Cloning Plan == | + | ==<b> Cloning Plan</b> == |
| - | Aim | + | <b>Aim</b> |
<br> | <br> | ||
# Build DiscoverY yeast: transform genes to S.pombe and turn it into the yeast we want. | # Build DiscoverY yeast: transform genes to S.pombe and turn it into the yeast we want. | ||
# BioBricks to submit: insert genes to vectors provided by iGEM (pSB1C3). | # BioBricks to submit: insert genes to vectors provided by iGEM (pSB1C3). | ||
<br> | <br> | ||
| - | Protocol | + | <b>Protocol</b> |
<br> | <br> | ||
# Design primer (1 day) – Team | # Design primer (1 day) – Team | ||
| Line 35: | Line 39: | ||
<br> | <br> | ||
| - | == PCR == | + | ==<b> PCR </b>== |
To manipulate gene products obtained from gene-bank, our team used PCR/Real-Time PCR methods. Commercially obtained genes contain few more base pairs added at front and back site. As for FGPR, the receptor protein of human has human-specific signal peptide at its starting point, and this region had to be replaced by S.pombe specific signal peptide to express the protein at membrane region. To do this, PCR primers containing signal peptide region of S.pombe were synthesized by method describeds below. Then, FGPR gene was amplified by PCR to get gene product whose signal region is replaced. Also, additional sequence at rear site of gene was removed. STAT gene went through similar process; removing additional region.<br> | To manipulate gene products obtained from gene-bank, our team used PCR/Real-Time PCR methods. Commercially obtained genes contain few more base pairs added at front and back site. As for FGPR, the receptor protein of human has human-specific signal peptide at its starting point, and this region had to be replaced by S.pombe specific signal peptide to express the protein at membrane region. To do this, PCR primers containing signal peptide region of S.pombe were synthesized by method describeds below. Then, FGPR gene was amplified by PCR to get gene product whose signal region is replaced. Also, additional sequence at rear site of gene was removed. STAT gene went through similar process; removing additional region.<br> | ||
<br> | <br> | ||
| - | + | <br> | |
| - | + | ||
<b>STAT</b><br> | <b>STAT</b><br> | ||
We need two kinds of brick to do PCR. One is STAT and the other is V_STAT. STAT would be cut by EcoRI and inserted into pBS1C3 for the submission. V_STAT would be cut by NdeI and BamHI sites and inserted into pREP41 vector for the expression<br> | We need two kinds of brick to do PCR. One is STAT and the other is V_STAT. STAT would be cut by EcoRI and inserted into pBS1C3 for the submission. V_STAT would be cut by NdeI and BamHI sites and inserted into pREP41 vector for the expression<br> | ||
| Line 60: | Line 63: | ||
<br> | <br> | ||
| - | == Oligo Synthesis == | + | ==<b> Oligo Synthesis </b>== |
Oligo-synthesis was supported by Bioneer.<br> | Oligo-synthesis was supported by Bioneer.<br> | ||
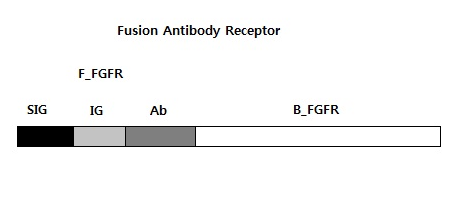
We requested synthesis of <b>fusion antibody gene</b> that is mainly used in our project.<br> | We requested synthesis of <b>fusion antibody gene</b> that is mainly used in our project.<br> | ||
| Line 81: | Line 84: | ||
<br><br><br> | <br><br><br> | ||
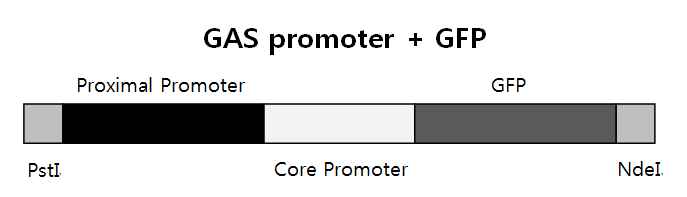
| - | == Homologous Recombination == | + | ==<b> Homologous Recombination</b> == |
Homologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks. Homologous recombination also produces new combinations of DNA sequences during meiosis, the process performed by many eukaryotes like animals and plants: production of sperm and egg cells. These new combinations of DNA promote genetic variations in offspring, which in turn enable populations to evolve. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. | Homologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks. Homologous recombination also produces new combinations of DNA sequences during meiosis, the process performed by many eukaryotes like animals and plants: production of sperm and egg cells. These new combinations of DNA promote genetic variations in offspring, which in turn enable populations to evolve. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. | ||
<br> | <br> | ||
| Line 92: | Line 95: | ||
<br> | <br> | ||
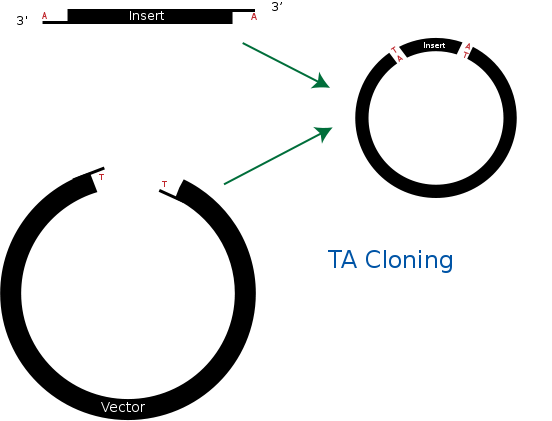
| - | == TA cloning == | + | == <b>TA cloning</b> == |
TA cloning is one of the subcloning techniques. This technique starts from product of PCR that uses taq DNA polymerase. Taq DNA polymerase tends to add one adenine overhang to the 3` end of PCR product. This result occurs because there is lack of ability to proofread from 3` to 5`. Therefore after PCR process, we can get amplified DNA sequences that has single adenine overhang on 3` end.<br> | TA cloning is one of the subcloning techniques. This technique starts from product of PCR that uses taq DNA polymerase. Taq DNA polymerase tends to add one adenine overhang to the 3` end of PCR product. This result occurs because there is lack of ability to proofread from 3` to 5`. Therefore after PCR process, we can get amplified DNA sequences that has single adenine overhang on 3` end.<br> | ||
In a mean time, it needs target vector for TA cloning. Target vector can be prepared by cutting target vector with blunt-end restriction enzyme. After this process, it is tailed with ddTTP using terminal transferase. Then target vector gets single thymine on each 3` ends. Finally, adenine on PCR product and thymine on target vector can bind together. This way, we can ligate PCR product and vector together without restriction enzyme.<br> | In a mean time, it needs target vector for TA cloning. Target vector can be prepared by cutting target vector with blunt-end restriction enzyme. After this process, it is tailed with ddTTP using terminal transferase. Then target vector gets single thymine on each 3` ends. Finally, adenine on PCR product and thymine on target vector can bind together. This way, we can ligate PCR product and vector together without restriction enzyme.<br> | ||
| Line 99: | Line 102: | ||
<br><br><br> | <br><br><br> | ||
| - | == References == | + | == <b>References</b> == |
Bioneer PCR service: <br> | Bioneer PCR service: <br> | ||
Latest revision as of 09:00, 13 October 2010
 "
"