Team:Imperial College London/Modelling
From 2010.igem.org
(Difference between revisions)
(Quick overview of protein display model ready) |
(Making picture area background of different colour) |
||
| Line 26: | Line 26: | ||
<li>Split TEV protease (<u>orange on diagrams below</u>), is an inactive, split form of TEV mounted on coiled coils. It can be activated again by coiled coils being cleaved by another active TEV.</li> | <li>Split TEV protease (<u>orange on diagrams below</u>), is an inactive, split form of TEV mounted on coiled coils. It can be activated again by coiled coils being cleaved by another active TEV.</li> | ||
</ol> | </ol> | ||
| - | {| style="width:850px;background:# | + | {| style="width:850px;background:#e7e7e7;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="5"; |
|- | |- | ||
|<html> | |<html> | ||
| - | <img src="http://www.openwetware.org/images/7/7f/Simple_production.JPG" width=" | + | <img src="http://www.openwetware.org/images/7/7f/Simple_production.JPG" width="280" alt="Diagram of simple production of Dioxygenase" /> |
</html> | </html> | ||
|<html> | |<html> | ||
| - | <img src="http://www.openwetware.org/images/1/1c/1-step_amplification.JPG" width=" | + | <img src="http://www.openwetware.org/images/1/1c/1-step_amplification.JPG" width="280" alt="Diagram of simple production of Dioxygenase" /> |
</html> | </html> | ||
|<html> | |<html> | ||
| - | <img src="http://www.openwetware.org/images/0/02/2-step_amplification.JPG" width=" | + | <img src="http://www.openwetware.org/images/0/02/2-step_amplification.JPG" width="280" alt="Diagram of simple production of Dioxygenase" /> |
</html> | </html> | ||
|- | |- | ||
| Line 68: | Line 68: | ||
<li>Receptor activation threshold was defined by 1 specific value as opposed to considering intermediate states between fully "off" and "on"</li> | <li>Receptor activation threshold was defined by 1 specific value as opposed to considering intermediate states between fully "off" and "on"</li> | ||
</ol> | </ol> | ||
| + | |||
| + | |||
|} | |} | ||
Revision as of 22:30, 11 October 2010
| Introduction to modelling |
In the process of designing our construct two major questions arose which could be answered by computer modelling:
|
| Quick overview of models | ||||||
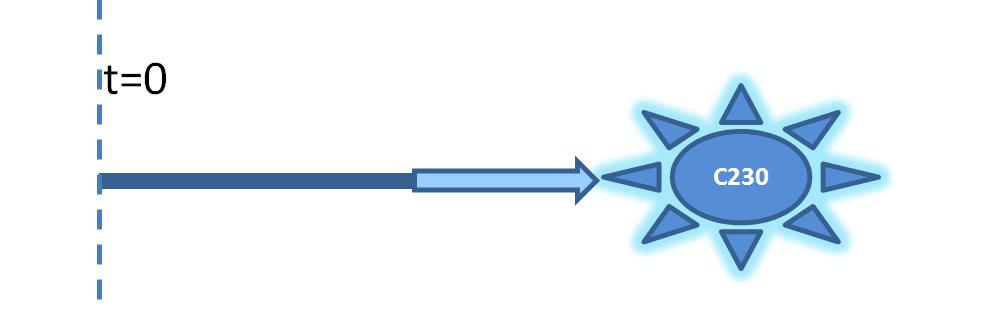
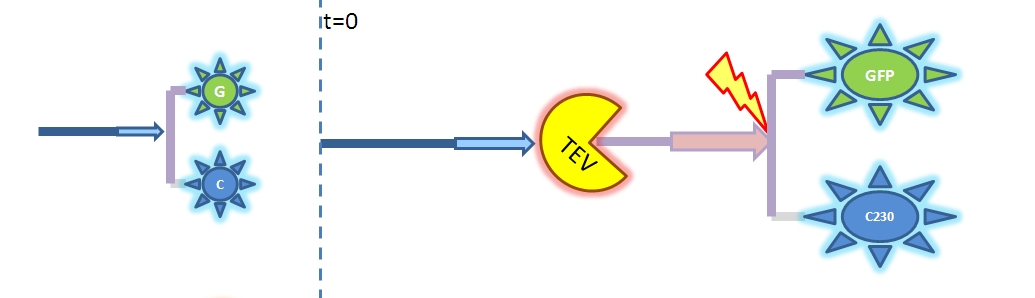
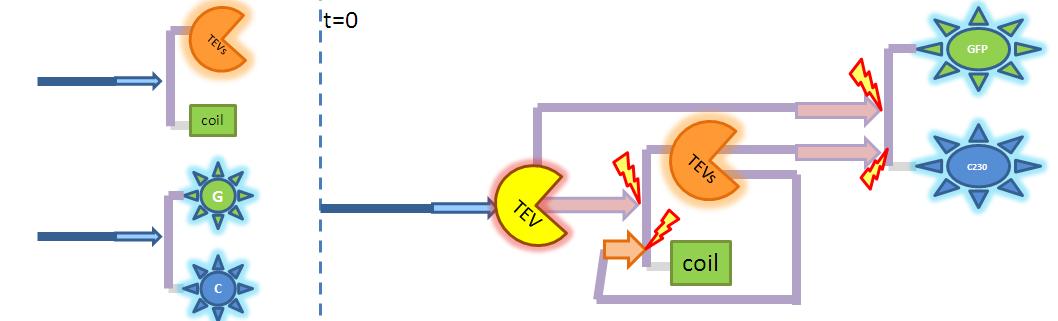
| Output Amplification Model Goals: This model was mainly developed in order to determine whether simple production is better than 1- or 2-step amplification. Further goals, contained estimation of the speed of modelled response.Elements of the system:
Major assumptions:
The aim of this model is to determine the concentration of Schistosoma elastase or TEV protease that should be added to bacteria to trigger the response. It is also attempted to model how long it takes for the protease or elastase to cleave enough peptides. Elements of the system:
Major assumptions:
|
| Results & Conclusions |
 "
"