Team:Calgary/8 June 2010
From 2010.igem.org
| Line 1: | Line 1: | ||
'''Tuesday June 8, 2010''' | '''Tuesday June 8, 2010''' | ||
| - | |||
| - | |||
| - | |||
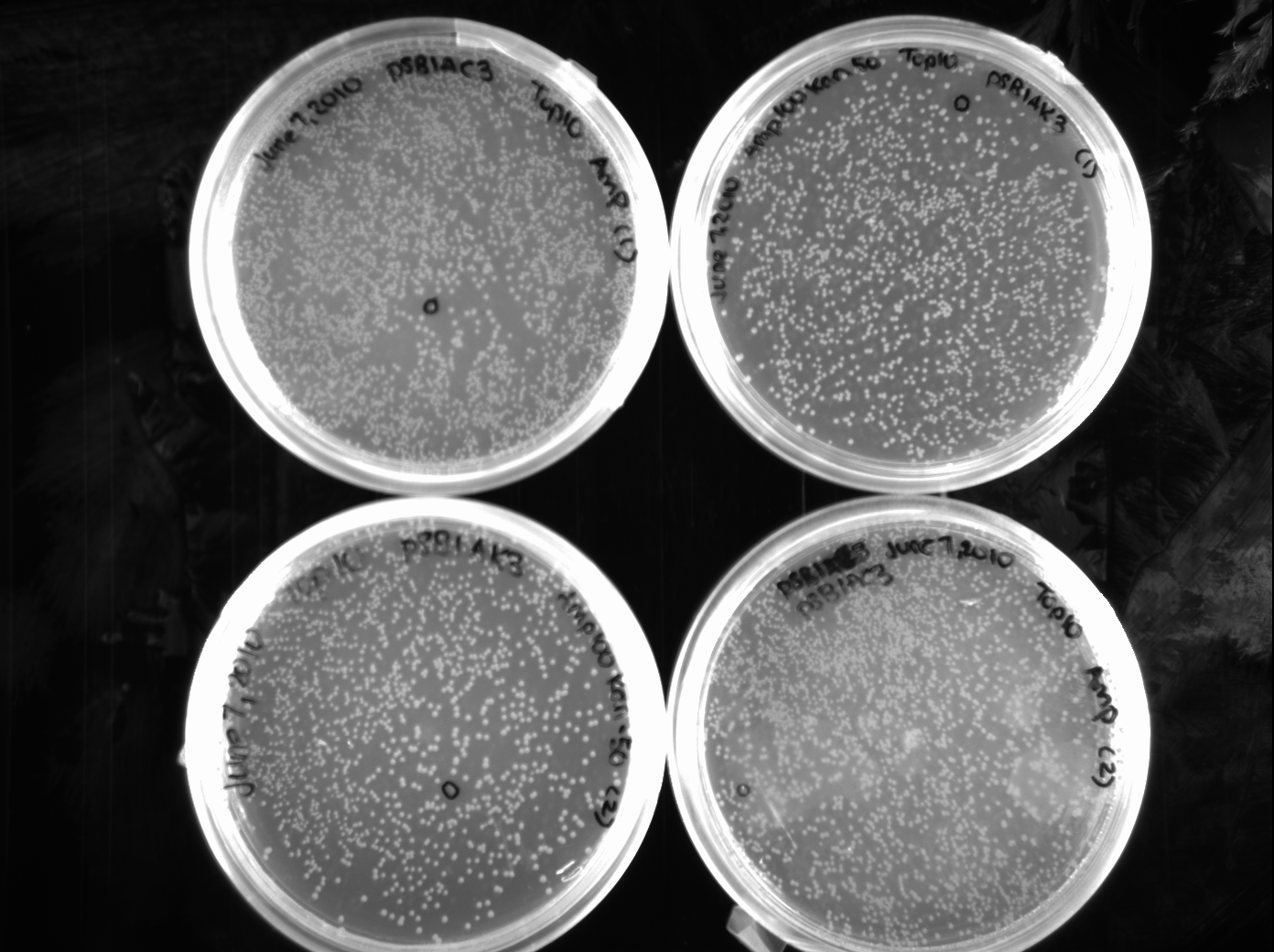
[[Image:06.08.2010.AC_and_AK_Plasmid_backbones.png|350px|thumb|Dev, Jeremy, and Chris's spread plates of Ampicillin - Chloramphenicol and Ampicillin - Kanamycin from the Parts Registry.]] | [[Image:06.08.2010.AC_and_AK_Plasmid_backbones.png|350px|thumb|Dev, Jeremy, and Chris's spread plates of Ampicillin - Chloramphenicol and Ampicillin - Kanamycin from the Parts Registry.]] | ||
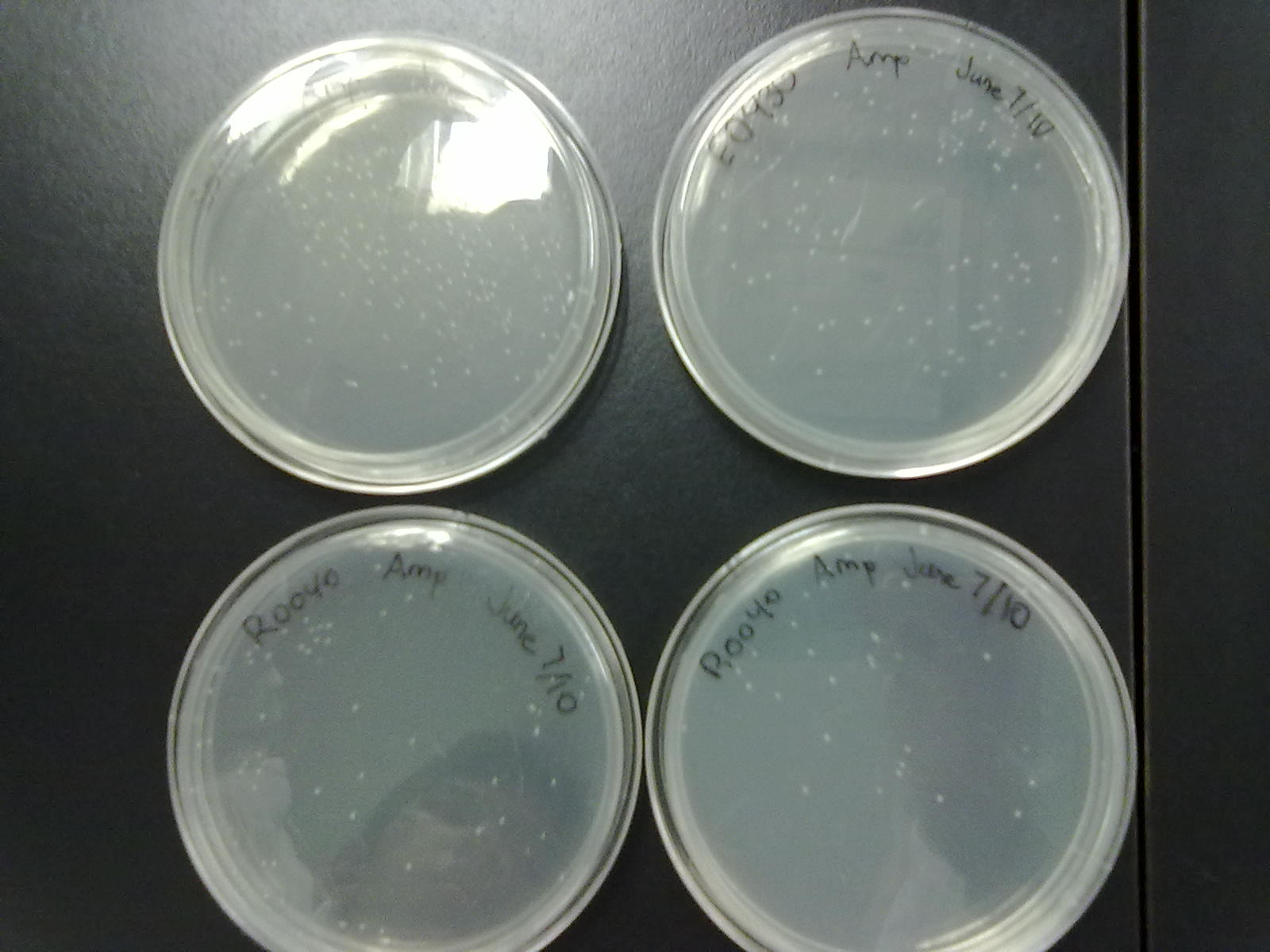
[[Image:06.08.2010.Amp_%2B_Kan_Plasmid_backbones.png|350px|thumb|Dev, Jeremy, and Chris's spread plates of the Ampicillin and Kanamycin plasmid backbones from the Parts Registry.]] | [[Image:06.08.2010.Amp_%2B_Kan_Plasmid_backbones.png|350px|thumb|Dev, Jeremy, and Chris's spread plates of the Ampicillin and Kanamycin plasmid backbones from the Parts Registry.]] | ||
| + | |||
| + | Today, we spent most of the day making overnight cultures from the transformed plates that were made yesterday. As a group, we began getting fundraising underway by finding contacts, dividng them among each person and setting a tentative goal of $20 000 to fundraise from corporate sponsors. We also organized how the money given from Alberta Innovates would be used | ||
| + | |||
| + | Chris, Jeremy, and Dev: Most of the growth on the plates were red, indicating that the placeholder gene was being transcribed and successfully produced. Our group spent today making overnight cultures so plasmid transfers could be done tomorrow. We made the cultures for plasmids pSB1A3, pSB1K2, pSB1AC3 and pSB1AK3 as well as restreaking the colonies that were used to innoculate the overnight cultures. Jeremy also transformed I732005 (lacZ gene for B-galactosidase) into competent cells so they could be used. | ||
Alex and Patrick: The ampicillin spread plates of the R0040 and E0430 parts grew successfully. We prepared an overnight culture of two colonies from each plate, totalling 8 Falcon tubes. Each colony innoculated was also restreaked on new ampicillin plates in the event that we require them again. We also helped Himika help Emily miniprep her parts. | Alex and Patrick: The ampicillin spread plates of the R0040 and E0430 parts grew successfully. We prepared an overnight culture of two colonies from each plate, totalling 8 Falcon tubes. Each colony innoculated was also restreaked on new ampicillin plates in the event that we require them again. We also helped Himika help Emily miniprep her parts. | ||
Revision as of 15:20, 15 June 2010
Tuesday June 8, 2010
Today, we spent most of the day making overnight cultures from the transformed plates that were made yesterday. As a group, we began getting fundraising underway by finding contacts, dividng them among each person and setting a tentative goal of $20 000 to fundraise from corporate sponsors. We also organized how the money given from Alberta Innovates would be used
Chris, Jeremy, and Dev: Most of the growth on the plates were red, indicating that the placeholder gene was being transcribed and successfully produced. Our group spent today making overnight cultures so plasmid transfers could be done tomorrow. We made the cultures for plasmids pSB1A3, pSB1K2, pSB1AC3 and pSB1AK3 as well as restreaking the colonies that were used to innoculate the overnight cultures. Jeremy also transformed I732005 (lacZ gene for B-galactosidase) into competent cells so they could be used.
Alex and Patrick: The ampicillin spread plates of the R0040 and E0430 parts grew successfully. We prepared an overnight culture of two colonies from each plate, totalling 8 Falcon tubes. Each colony innoculated was also restreaked on new ampicillin plates in the event that we require them again. We also helped Himika help Emily miniprep her parts.
Himika: Himika made overnight cultures from B0034 (the RBS that we all need) and B0015 (the double terminator for the end of all our circuits).
Emily: Today I did gel electrophoresis of yesterday’s Colony PCR. I also did a miniprep of yesterday’s overnight cultures of both I0500 and I13453 and I determined the concentration using the Nanodrop spectrophotometer. I then performed a restriction digest with EcoRI and PstI. This is another verification step for the arabinose promoters. If it works, we would expect to see band sizes at ~ 1.4 kb (1210 bp for the gene + 200 bp for the Biobrick Prefix and Suffix) for I0500 as well as bands at ~ 330 bp (230 bp for the gene + 200 bp for the Biobrick Prefix and Suffix). I also spent some time looking into what kind of genes of interest we could use to test out our system. I’ve been looking into some human genes that have been found to misfold in E Coli. These could work as a positive control, in order to demonstrate that our system is working.
 "
"