Team:Stockholm/10 September 2010
From 2010.igem.org
(→Andreas) |
|||
| Line 80: | Line 80: | ||
Plate grown ON in 37 °C. | Plate grown ON in 37 °C. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === Restriction enzymes control === | ||
| + | |||
| + | ==== Digestion ==== | ||
| + | |||
| + | {| | ||
| + | ! mix | ||
| + | | (µl) | ||
| + | | rowspan="6" width="100" | | ||
| + | ! colspan="2" | Conditions | ||
| + | |- | ||
| + | | sH<sub>2</sub>O | ||
| + | | 15 | ||
| + | ! time | ||
| + | ! °C | ||
| + | |- | ||
| + | | 10x buffer | ||
| + | | 3 | ||
| + | | 30m | ||
| + | | 37 | ||
| + | |- | ||
| + | | DNA (pSB1C3) | ||
| + | | 10 | ||
| + | | 20m | ||
| + | | 65 | ||
| + | |- | ||
| + | | enzyme (E/S) | ||
| + | | 1 | ||
| + | | OO | ||
| + | | 10 | ||
| + | | align="right" | tot | ||
| + | | 29µl | ||
| + | | colspan="2" | | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==== Gel ==== | ||
| + | |||
| + | [[Image:place_for_picture.jpg|200px|thumb|left|]] | ||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | pSB1C3 cut E | ||
| + | |- | ||
| + | | 3 | ||
| + | | pSB1C3 cut S | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | === MITF-M === | ||
| + | |||
| + | ==== Site-Directed Mutagenesis ==== | ||
| + | |||
| + | *Add Dpn1 (12.30-16=3.5h) | ||
| + | *Deactivate Dpn1 at 80°C for 20m | ||
Revision as of 12:30, 4 October 2010
Contents |
Andreas
Cloning of N-CPPs into pSB1C3 & Extraction of RBS BioBrick (BBa_B0030)
Transformations from 9/9 resulted in good colony yields on all plates. Chose "pSB1C3.N-CPP* 9/9" for colony PCR.
Colony PCR
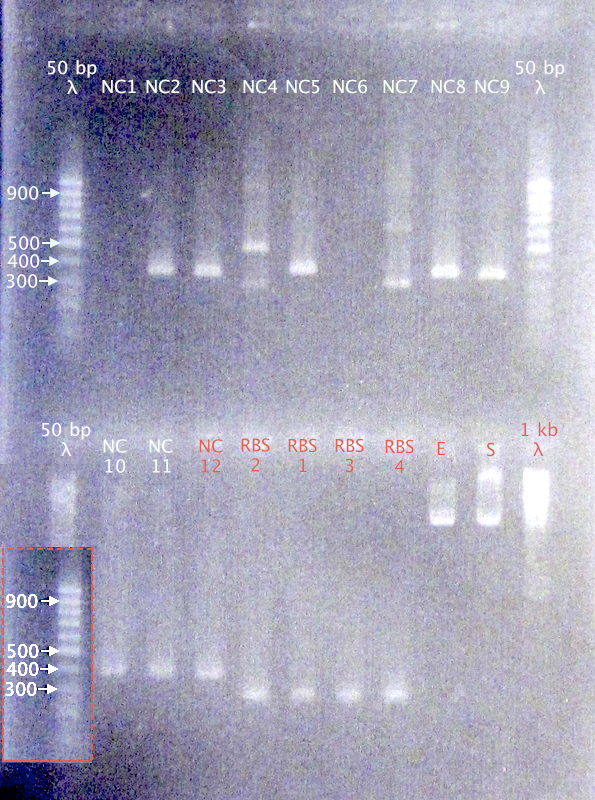
Picked 12 N-CPP* clones (NC 1-12) and 4 RBS clones (RBS 1-4)
| PCR tubes | |
|---|---|
| dH2O | 16.22 |
| DreamTaq buffer | 2 |
| dNTPs, 10 mM | 0.4 |
| VF2 | 0.4 |
| VR | 0.4 |
| DreamTaq pol. | 0.08 |
| Template DNA | 0.5 |
| 20 μl | |
PCR settings
Standard colony PCR protocol.
- 1:00 elongation
Gel verification
Also ran two samples for Mimmi (E & S)
1.5 % agarose, 90 V
Expected bands
- N-Tra10: 389 bp
- N-TAT: 359 bp
- N-LMWP: 368 bp
- RBS B0030: 253 bp
Results
- N-CPPs: Potentially correct bands for clones 2, 3, 5, 8, 9, 10, 11 & 12
- RBS B0030: Correct-sized bands for all four clones.
ON cultures
Set ON cultures for all relevant N-CPPs, for plasmid prep.
- N-CPP 2, 3, 5, 8, 9, 10, 11, 12
- 5 ml LB + 25 Cm
- 37 °C, 220 rpm
Selected clone 4 of RBS 30. Both plasmid prep and glycerol stock.
- RBS 30 4
- 5 ml LB + 100 Amp
- 37 °C, 220 rpm.
- 3 ml LB + 100 Amp
- 30 °C
- 5 ml LB + 100 Amp
Extraction of RBS BioBrick (BBa_B0034)
After studying the original RBS of our expression vector pEX, we decided that BBa_B0034 was a better candidate for our SOD/yCCS operon than BBa_B0030, as it better resembles the RBS of pEX, as well as minimizes the distance from the first gene in the operon.
Extracted BBa_B0034 (RBS 34), carried on pSB1A2, from iGEM plate 1, well 2M. Transformed into Top10.
- Quick transformation
- 1 μl DNA
- Amp 100
Preparation of chemically competent Top10
Since I've previously experienced slight AmpR contamination in our latest batch of competent Top10, I streaked an LB agar plate with competent Top10 cells to isolate single clones.
Plate grown ON in 37 °C.
Mimmi
Restriction enzymes control
Digestion
| mix | (µl) | Conditions | |||||
|---|---|---|---|---|---|---|---|
| sH2O | 15 | time | °C | ||||
| 10x buffer | 3 | 30m | 37 | ||||
| DNA (pSB1C3) | 10 | 20m | 65 | ||||
| enzyme (E/S) | 1 | OO | 10 | tot | 29µl | ||
Gel
| well | sample |
|---|---|
| 1 | ladder |
| 2 | pSB1C3 cut E |
| 3 | pSB1C3 cut S |
MITF-M
Site-Directed Mutagenesis
- Add Dpn1 (12.30-16=3.5h)
- Deactivate Dpn1 at 80°C for 20m
 "
"