Team:ETHZ Basel/Modeling/Interworking
From 2010.igem.org
| Line 22: | Line 22: | ||
Because of the limited amount of different combination possibilities (48), it was chosen to evaluate all combinations. Parameter space of variable experimental implementation possibilities is shown in Table 1. | Because of the limited amount of different combination possibilities (48), it was chosen to evaluate all combinations. Parameter space of variable experimental implementation possibilities is shown in Table 1. | ||
| - | {| border="1" | + | {| border="1" align="center" |
|+ '''Table 1: Experimental parameters''' | |+ '''Table 1: Experimental parameters''' | ||
! Che? !! LSP? !! LSP?' !! [Asp] !! [AP] !! [anchor] | ! Che? !! LSP? !! LSP?' !! [Asp] !! [AP] !! [anchor] | ||
Revision as of 08:55, 27 September 2010
Evaluation for wet laboratory
Goals of the evaluation
To determine the best possible parts for the biological implementation of E. lemming, the combined molecular model (Light switch - Chemotaxis) has been used to answer specific questions:
- Which receptor (Che) protein should be attacked?
- To which light-sensitive protein (PhyB, PIF3) should the receptor protein be linked?
- What is a good aspartate concentration?
- In what quantity should the anchor and binding light-sensitive protein be chosen?
Answers to this questions were able to decrease the experimental expenses significantly.
Evaluator
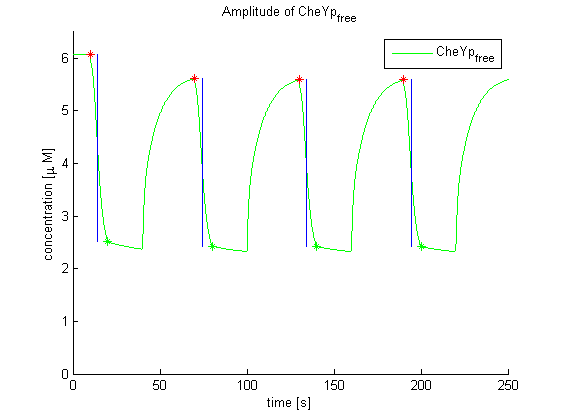
Evaluation of the results is a general optimization problem. The output variable in the case of E. lemming is the concentration of free and phosphorylated CheYp, because it is directly linked to the movement bias. To achieve a reasonable evaluation variable, the relative amplitude of free and phosphorylated CheYp was chosen to be maximized.
Because of the limited amount of different combination possibilities (48), it was chosen to evaluate all combinations. Parameter space of variable experimental implementation possibilities is shown in Table 1.
| Che? | LSP? | LSP?' | [Asp] | [AP] | [anchor] |
|---|---|---|---|---|---|
| CheR | PhyB | PIF3 | 0 uM | 0 uM | 0 uM |
| CheB | PIF3 | PhyB | 10^-6 uM | 25 uM | 25 uM |
| CheY | 10^-3 uM | 50 uM | 50 uM | ||
| CheZ | 75 uM | 75 uM | |||
| 100 uM | 100 uM |
Results
Evaluation for information processing
Goal of the evaluation
Main goal of the evaluation for information processing was to determine time constants of E. lemming by using the combined molecular model (Light switch - Chemotaxis).
 "
"