Team:Cambridge/Gibson/Mechanism
From 2010.igem.org
(→Gibson Assembly) |
(→Gibson Assembly) |
||
| Line 24: | Line 24: | ||
<html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/b/bf/Cambridge-Gib3.png" style="border:1px solid black"></div></html> | <html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/b/bf/Cambridge-Gib3.png" style="border:1px solid black"></div></html> | ||
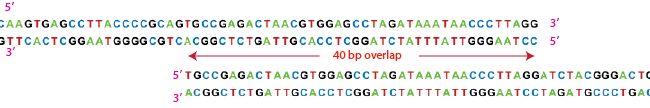
| + | Once it has chewed back far enough A-T G-C base pairing allows the two pieces to bind together. | ||
<html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/5/57/Cambridge-Gib5.png" style="border:1px solid black"></div></html> | <html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/5/57/Cambridge-Gib5.png" style="border:1px solid black"></div></html> | ||
| - | |||
<html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/1/1f/Cambridge-Gib6.png" style="border:1px solid black"></div></html> | <html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/1/1f/Cambridge-Gib6.png" style="border:1px solid black"></div></html> | ||
| + | |||
| + | We now have a single piece of DNA but it is not physically ligated together, it is merely held together by hydrogen bonding, also there are gaps in both single strands. | ||
<html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/8/8f/Cambridge-Gib7.png" style="border:1px solid black"></div></html> | <html><div style="text-align:center"><img src="https://static.igem.org/mediawiki/2010/8/8f/Cambridge-Gib7.png" style="border:1px solid black"></div></html> | ||
Revision as of 17:45, 18 September 2010

Gibson Assembly is a means to join overlapping DNA sequences, technically it does not describe the way in which these sequences are created. However since this will be of importance to iGEM teams, we will briefly discuss this.
Creating overlapping DNA sequences
Overlapping DNA sequences can be created by PCR. We can add twenty base-pairs to the end of a sequence by using a primer which runs as follows from 5' to 3'.
20 bp of sequence to add -> 20 bp of template to anneal to.
By using two such primers we can add 20 bp of sequence A to sequence B and 20 bp of sequence B to sequence A. We are then ready to use Gibson Assembly.
Gibson Assembly
Gibson Assembly mix contains 3 enzymes:
- T5 exonuclease
- Phusion polymerase
- Taq ligase
The Gibson reaction relies on the action of the T5 exonuclease - this chews back at the 5' ends of both pieces of DNA


Once it has chewed back far enough A-T G-C base pairing allows the two pieces to bind together.


We now have a single piece of DNA but it is not physically ligated together, it is merely held together by hydrogen bonding, also there are gaps in both single strands.











 "
"