Team:Stockholm/16 September 2010
From 2010.igem.org
(Difference between revisions)
(New page: {{Stockholm/Top2}} ==Andreas==) |
(→Andreas) |
||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
==Andreas== | ==Andreas== | ||
| + | ===Assembly of new parts=== | ||
| + | |||
| + | ====Gel verification of part digestions==== | ||
| + | Ran gels of digestion samples in parallel with undigested samples to verify successful digestions and insert sizes. | ||
| + | |||
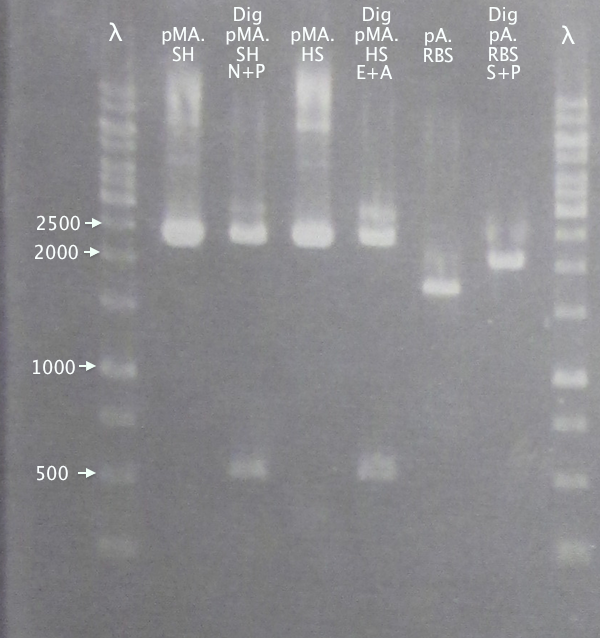
| + | '''Gel 1'''<br /> | ||
| + | [[image:Gelver_diggel1_16sep.png|200px|thumb|right|'''Gel verification of digested samples, gel 1.''' <br />&4 μl λ; 3 μl sample.<br />λ = O'GeneRuler 1 kb DNA ladder.]] | ||
| + | 1 % agarose, 110 V | ||
| + | |||
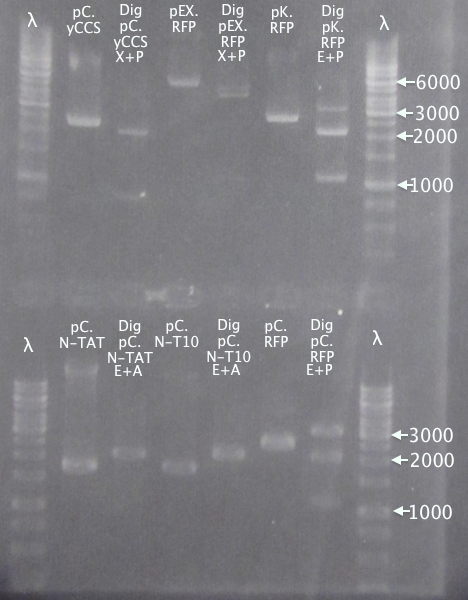
| + | '''Gel 2'''<br /> | ||
| + | [[image:Gelver_diggel2-3_16sep.png|200px|thumb|right|'''Gel verification of digested samples, gel 2.''' <br />&4 μl λ; 3 μl sample.<br />λ = O'GeneRuler 1 kb DNA ladder.]] | ||
| + | 1 % agarose, 110 V | ||
| + | |||
| + | '''Results'''<br /> | ||
| + | Successful digestion with corresponding bands for all digested samples. N-TAT and N-Tra10 not verified, since gel was run too far, but plasmid linearization should indicate successful digestion. | ||
| + | |||
| + | Most samples show somewhat incomplete digestion. For digestions with FastDigest enzymes, this may be an indication of old/inactive enzymes. Especially PstI should be analyzed for activity. | ||
| + | |||
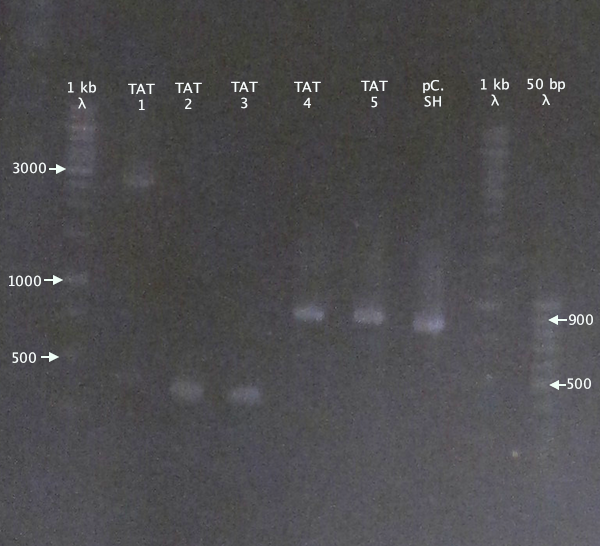
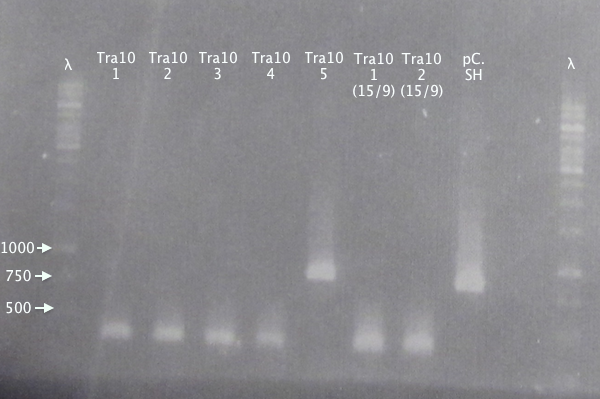
| + | ====Colony PCR==== | ||
| + | Picked new colonies for colony PCR from 14/9 plates: | ||
| + | *pSB1K3.TAT⋅SOD⋅His: TAT⋅SH 1-5 | ||
| + | *pSB1K3.Tra10⋅SOD⋅His: T10⋅SH 1-5 | ||
| + | *pEX.SOD 1-4 | ||
| + | |||
| + | Standard colony PCR settings. | ||
| + | *Elongation: 1:30 | ||
| + | |||
| + | ====Gel verification==== | ||
| + | [[image:ColPCR_TAT*SH_16sep.png|200px|thumb|right|'''Colony PCR gel verification of N-TAT⋅SOD⋅His clones.'''<br />4 μl λ; 5 μl sample.<br />1 kb λ = O'GeneRuler 1 kb DNA ladder; 50 bp λ = GeneRuler 50 bp DNA ladder.]] | ||
| + | [[image:ColPCR_Tra10*SH_16sep.png|200px|thumb|right|'''Colony PCR gel verification of N-Tra10⋅SOD⋅His clones.'''<br />4 μl λ; 5 μl sample.<br />λ = O'GeneRuler 1 kb DNA ladder.]] | ||
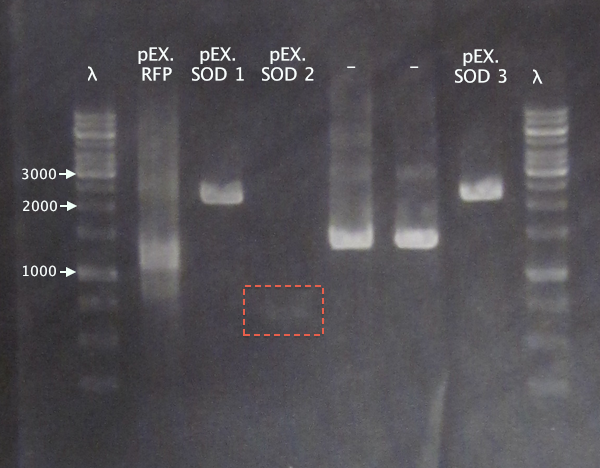
| + | [[image:ColPCR_pEX.SOD_16sep.png|200px|thumb|right|'''Colony PCR gel verification of pEX.SOD clones.'''<br />4 μl λ; 5 μl sample.<br />λ = O'GeneRuler 1 kb DNA ladder.]] | ||
Revision as of 19:06, 16 September 2010
Contents |
Andreas
Assembly of new parts
Gel verification of part digestions
Ran gels of digestion samples in parallel with undigested samples to verify successful digestions and insert sizes.
Gel 1
1 % agarose, 110 V
Gel 2
1 % agarose, 110 V
Results
Successful digestion with corresponding bands for all digested samples. N-TAT and N-Tra10 not verified, since gel was run too far, but plasmid linearization should indicate successful digestion.
Most samples show somewhat incomplete digestion. For digestions with FastDigest enzymes, this may be an indication of old/inactive enzymes. Especially PstI should be analyzed for activity.
Colony PCR
Picked new colonies for colony PCR from 14/9 plates:
- pSB1K3.TAT⋅SOD⋅His: TAT⋅SH 1-5
- pSB1K3.Tra10⋅SOD⋅His: T10⋅SH 1-5
- pEX.SOD 1-4
Standard colony PCR settings.
- Elongation: 1:30
 "
"