Team:Stanford/Notebook/Lab Work/Week 3
From 2010.igem.org
(→7/6 Tuesday) |
(→7/16 Friday) |
||
| (101 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template: | + | {{Template:Stanford_2010_Main_Menu}} |
| - | {{Template: | + | {{Template: Stanford_2010_Notebook}} |
<div style="width:720px;text-align:justify"> | <div style="width:720px;text-align:justify"> | ||
<!-- start writing here --> | <!-- start writing here --> | ||
| - | ==7/ | + | ==7/12 Monday== |
| - | + | ||
| - | + | ===Planning Meeting Notes=== | |
| - | * | + | *Leader: Alex |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Alex's Notebook=== | |
| - | + | Run three gels: | |
| - | + | 4A5 (3395- 1069) w/ 2K3 (4425-1069) | |
| + | F2620 (1061-2079) w/ I0500 (1210-4425) | ||
| + | I8 (2093-2079=>936 and 1143) w/ T9 (1945-2157=> 936 and 1221). | ||
| + | Gel extract. | ||
| + | Run diagnostic. | ||
| + | Ligate. | ||
| - | |||
| - | |||
| - | + | Minipreped inoculations. | |
| - | + | 3C5: 123.5 ng/mL 280: 1.90 230: 2.86 | |
| + | 1K3: 84 ng/mL 280: 1.86 230: 2.58 | ||
| + | I0500: 54.7 ng/mL 280: 1.90 230: 2.32 | ||
| + | I746908: 234.3 ng/mL 280: 1.85 230: 1.90 | ||
| + | |||
| + | Ligation/Promoter characterization (2 10X buffer, 1 ligase): | ||
| - | + | DNA vector 250-500 ng 400 ng | |
| - | + | Insert 5-20x of vector 6 ug | |
| - | + | Endy’s Promoter | |
| - | + | F2620 + Superfolded GFP (PCR) + pSB1A2 | |
| - | + | F2620 + Superfolded GFP (PCR) + pSB3C5 | |
| - | + | Bad Promoter | |
| - | + | Testing of I746908 (already in pSB1A2) | |
| + | I746908 + pSB3C5 (10, 5) | ||
| - | + | Endy’s Promoter | |
| + | T9002 + 1A2 (10, 5) | ||
| + | T9002 + 3C5 (10, 5) | ||
| + | Bad Promoter | ||
| + | I0500 + E0240 + pSB1A2 (5, 5, 5) | ||
| + | I0500 + E0240 + pSB3C5 (5, 5, 5) | ||
| - | + | Promoter/primer design | |
| - | + | 20 bp | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ===Christopher's Notebook=== |
| - | + | *Run Gel of Restricted Digested RBS+GFP+Terminators (I746908 partial) | |
| - | * | + | *Gel Extract |
| - | * | + | *Run Gel of Gel Extracted Digest Products from Friday: |
| + | 3C5 E/P 2738 bp (buffer 3) | ||
| + | F2620 E/S (on pSB1A2) 1061 bp | ||
| + | *I746908 E/P (on psb1A2) 2093 bp-can’t separate | ||
| + | *B0034 X/P (on psb1A2) 12 bp-do NOT run on gel | ||
| + | *J23100 E/S (on psb1A2) 35 bp-do NOT run on gel | ||
| - | + | === Karina's Notebook === | |
| + | |||
| + | Goal: We couldn't extract the terminators because they were too small and ran off the gel. After discussing with Laura and Francisco, we decided to clone GFP and RFP into the plasmid in which the terminators come (pSB1AK3). <br/><br/> | ||
| + | |||
| + | '''Make BSA''' | ||
| + | *BSA comes as 100x | ||
| + | *made 1 mL of 1x solution by adding 900uL H20 and 100 uL BSA <br/><br/> | ||
| + | |||
| + | '''Cut the GFP and RFP linearized plasmid (prev. cut at S and P) at EcoRI restriction site ''' | ||
| + | *used recipe as described on [[Team:Stanford/Notebook/Lab_Work/Week_2 | July 9, 2010]] | ||
| + | *only used 1 enzyme, therefore used 27 uL H20 | ||
| + | *put in waterbath at 10:30 am, and plan to take out after lunch. <br/><br/> | ||
| + | |||
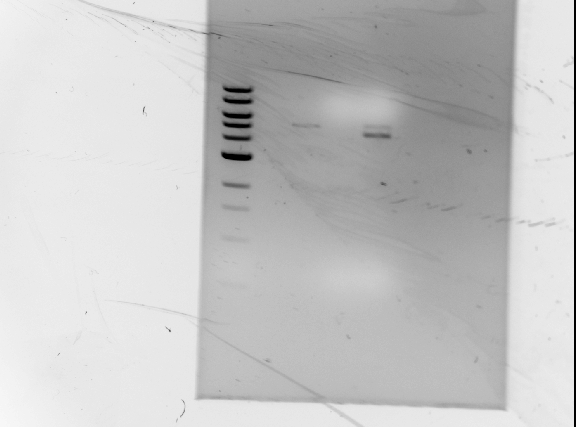
| + | '''Results of diagnostic gel''' <br/> | ||
| + | [[Image:071210_gel.jpg | none | frame | left to right: 1 kb ladder, GFP plasmid (cut at S, P), | ||
| + | RFP plasmid (cut at S, P), GFP plasmid (cut at S, P, and cut again at E), RFP plasmid (cut at S, P, and cut again at E) | ||
| + | Note: GFP lanes have no visible bands]]<br/> | ||
| + | |||
| + | ''Observations/Troubleshooting'':<br/> | ||
| + | *no DNA bands for GFP<br/> | ||
| + | ** not enough DNA?<br/> | ||
| + | ** will double DNA concentration during digests<br/> | ||
| + | *bands are too large to be RFP | ||
| + | **possible partial digest? | ||
| + | **leave digests overnight | ||
| + | <br/> | ||
| + | |||
| + | '''Digests'''<br/> | ||
| + | Digest miniprepped GFP, RFP and Terminators.<br/> | ||
| + | *Cut GFP and RFP at E and S | ||
| + | *Cut terminators at E and X<br/><br/> | ||
| + | ''Recipe''<br/> | ||
| + | *25 uL DNA<br/> | ||
| + | *1.0 uL enzyme 1<br/> | ||
| + | *1.0 uL enzyme 2<br/> | ||
| + | *5.0 uL NEB Buffer 2<br/> | ||
| + | *5.0 uL BSA (10x) <br/> | ||
| + | *13 uL H20<br/> | ||
| + | Total Volume: 50 uL<br/> | ||
| + | We used twice as much DNA than the usual protocol because our DNA concentrations from the minipreps were very low. <br/> | ||
| + | Prepared 3 reactions for each RSID; ran 6 PCR reactions total. | ||
| + | Ran digests overnight | ||
| + | |||
| + | ===Francisco's Notebook=== | ||
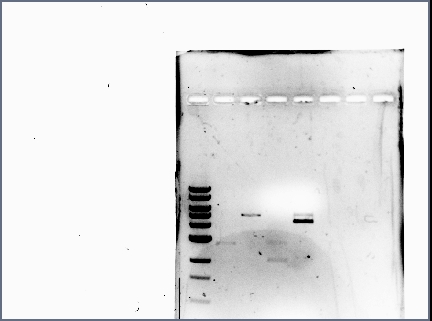
| + | *Reran the diagnostic gel (see Karina's Notebook): | ||
| + | |||
| + | [[Image:071210_gel_2.jpg | none | frame | left to right: 1 kb ladder, GFP and RFP plasmid (cut at S, P), GFP and RFP plasmid (cut at S, P, and also at E) | ||
| + | Note: GFP lanes now have (faintly) visible bands]] | ||
| + | |||
| + | *Lessons learned: | ||
| + | **When loading small volumes, use narrow wells. | ||
| + | **When suspecting there are no bands, increase the exposure time to confirm. | ||
| + | |||
| + | ===Greg's Notebook=== | ||
| + | |||
| + | *Ran gel with pSB3c5, F2620, I746908 | ||
| + | **I746908 didn't work, extracted pSB3c5 and F2620 | ||
| + | *Worked on MDV powerpoint presentation | ||
| + | *Ran diagnostic gel of gel extraction results | ||
| + | **pSB3c5 worked, F2620 left only a faint line | ||
| + | *With Alex, miniprepped pSB1k3, I746908, I0500 | ||
| + | *Performed nanodrop on miniprep: | ||
| + | {| | ||
| + | |part #|| 280 || 230 || concentration (ng/uL) | ||
|- | |- | ||
| - | + | |I0500r || 1.87 || 1.28 || 62.5 | |
|- | |- | ||
| - | | | + | |pSB1k3 || 1.81 || 1.14 || 67.6 |
|- | |- | ||
| - | | | + | |I746908 || 1.89 || 1.73 || 175.7 |
| + | |} | ||
| + | |||
| + | *Digested more F2620, pSB3c5, and I0500. Also digested 3 superfolded GFP inserts | ||
| + | |||
| + | ==7/13 Tuesday== | ||
| + | ===Alex's Notebook=== | ||
| + | *Streak bacteria | ||
| + | *Back inoculate bacteria 500 ul | ||
| + | |||
| + | |||
| + | ===Christopher's Notebook=== | ||
| + | *Run Gels (0.8%) of Restriction Digests: | ||
| + | 1M, 2M, 2J (X/P) | ||
| + | F2620 (E/S) | ||
| + | 3C5 (E/P) | ||
| + | I0500 (E/S) | ||
| + | |||
| + | '''Note: for PCR of I746908: after first gel of PCR product, gel extract, and then do a restriction digest. After restriction digest, do a reaction cleanup; pieces are ready for ligation'''. | ||
| + | |||
| + | ===Laura's Notebook=== | ||
| + | *restreaked plates from 7/1/10 (with Karina) | ||
| + | *for each plate, pick 8 colonies and streak, starting at the center of the "pie piece," and working towards the perimeter of the plate | ||
| + | *plate ID's: | ||
| + | |||
| + | ===Greg's Notebook=== | ||
| + | *Made gel, ran digests from yesterday | ||
| + | *Extracted pBAD (I0500) and pSB3c5 | ||
| + | *Practiced primer design | ||
| + | *Inventoried my box | ||
| + | |||
| + | ==7/14 Wednesday== | ||
| + | ===Alex's Notebook=== | ||
| + | *Order oligos/primers | ||
| + | *Ligate remaining sets | ||
| + | *Streak new bacteria | ||
| + | *Plate old bacteria | ||
| + | *PPT presentation | ||
| + | |||
| + | |||
| + | ===Laura's Notebook=== | ||
| + | *plates restreaked yesterday look good- bacteria grew well, some individual colonies | ||
| + | *helped Francisco with minipreps (Promega kit) of : terminator, GFP, RFP | ||
| + | **Francisco set up liquid cultures yesterday afternoon | ||
| + | *Francisco poured, ran 0.8% gel | ||
| + | **yesterday's digestions, with today's minipreps as controls | ||
| + | *gel extraction: (I cut out bands, Francisco and Karina extracted) | ||
| + | {| | ||
| + | |part || tube mass (g) || tube + gel (g) || gel (g) || volume QG (uL) | ||
|- | |- | ||
| - | | | + | |terminator || 1.03 || 1.14 || 0.11 || 330 |
|- | |- | ||
| - | | | + | |RFP || 1.11 || 1.18 || 0.07 || 210 |
|- | |- | ||
| - | | | + | |GFP || 1.03 || 1.13 || 0.10 || 300 |
| + | |} | ||
| + | |||
| + | ===Francisco's Notebook=== | ||
| + | *We've decided to cut out GFP and RFP inserts and ligate them into a linearized terminator plasmid. GFP and RFP inserts are fairly large (~700 bp) and would be easier to gel extract than a terminator insert. The terminator we are using is B1006. | ||
| + | |||
| + | *Mini-prepped GFP, RFP and terminator plasmids. Mini-prep results: | ||
| + | {| border = "1" | align = center | ||
| + | ! Part !! 260/280 !! 260/230 !! ng/uL | ||
|- | |- | ||
| - | | | + | | Term || 1.84 || 0.87 || 62.9 |
| + | |- | ||
| + | | RFP || 1.87 || 1.65 || 235.5 | ||
| + | |- | ||
| + | | GFP || 1.85 || 1.05 || 121.7 | ||
|} | |} | ||
| - | ==7/ | + | *Digested RFP and GFP plasmids with EcoR1 and Spe1; terminator plamids with EcoR1 and Xba1. |
| + | *Gel extracted the RFP and GFP inserts and the linearized terminator plasmid. | ||
| + | |||
| + | ===Greg's Notebook=== | ||
| + | |||
| + | *Worked on MDV presentation | ||
| + | |||
| + | |||
| + | |||
| + | ==7/15 Thursday== | ||
| + | ===Alex's Notebook=== | ||
| + | |||
| + | Check plates; streak new ones; back dilute for a third time. | ||
| + | Practice PPT for MDV | ||
| + | |||
| + | ==7/16 Friday== | ||
| + | ===Recap Meeting Notes=== | ||
| + | *No recap meeting- presented at MDV this morning. | ||
| + | |||
| + | |||
| + | ===Alex's Notebook=== | ||
| + | Due Monday morning: | ||
| + | Cloning scheme; check list on wiki for Christina and Drew. | ||
| + | Create a timeline for our own use. Deadlines as well. | ||
| + | |||
| + | ===Francisco's Notebook=== | ||
| + | |||
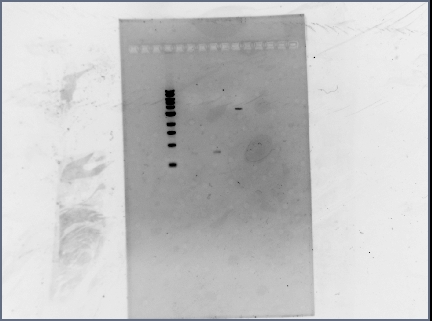
| + | *Ran a diagnostic gel of the digestion done on Wednesday | ||
| + | **GFP and RFP plasmids (cut at E, S); Term plasmid (cut at E, X): | ||
| + | |||
| + | [[Image: 071310_gel.jpg | none | frame | left to right: 1 kb ladder, RFP (cut at E, S), GFP (cut at E, S), Term (cut at E, X). Notes: Loaded every other lane, RFP lane has extremely faint band (but not visible in this image). Ran at 95V for 45 min.]] | ||
| + | |||
| + | *Results: the terminator backbone has a bright band, but the gel extracted GFP and RFP inserts had very faint bands (low concentrations). When we ligate, we may need to use larger volumes of the inserts. | ||
| + | |||
| + | ===Chris's Notebook=== | ||
| + | |||
| + | ====Ligations:==== | ||
| + | F2620+sfGFP+psb1A2 | ||
| + | pBAD+sfGFP+psb1A2 | ||
| + | F2620+sfGFP+psb3C5 | ||
| + | pBAD+sfGFP+psb3C5 | ||
| + | |||
| + | ====Ligation (20 μl reaction volume):==== | ||
| + | |||
| + | *DNA vector 250-500 ng (5 ul of vector) | ||
| + | *Insert 5-20x of vector (equal amounts 5 ul of each insert) | ||
| + | *10x buffer 2 μl | ||
| + | *Ligase 1 μl | ||
| + | *ATP (optional) 1 μl of 10 mM stock (4°C) | ||
| + | *NP water Bring to 20 μl total volume | ||
| + | |||
| + | Ligate at RT for 2 hr (sticky end) or 2 hr (blunt end) | ||
| + | Transform 0.5 and 1.0 ul | ||
| + | |||
| + | ==7/17 Saturday== | ||
| + | ===Alex's Notebook=== | ||
| + | |||
| + | Check plates again for no growth. | ||
| + | Restreaked again. | ||
| + | |||
| + | Run gels for restriction digest products; 120 ul each plasmid type, 240 total. | ||
| + | Ten wells minimum total, 4 more lanes for space. | ||
| + | Gel extract (partial). | ||
| + | Minipreped B0014. | ||
| + | |||
| + | |||
| + | ==7/18 Sunday== | ||
| + | ===Alex's Notebook=== | ||
| + | |||
| + | Gel extract (complete). | ||
| + | Streak, possible success. | ||
| + | Digested B0014 (X/P). | ||
| - | |||
</div> | </div> | ||
Latest revision as of 17:12, 14 September 2010

| Home | Project | Applications | Modeling | Parts | Team | Notebook |
Spring: Brainstorming | Spring Meetings
Summer: Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Summaries
|
7/12 Monday
Planning Meeting Notes
- Leader: Alex
Alex's Notebook
Run three gels: 4A5 (3395- 1069) w/ 2K3 (4425-1069) F2620 (1061-2079) w/ I0500 (1210-4425) I8 (2093-2079=>936 and 1143) w/ T9 (1945-2157=> 936 and 1221). Gel extract. Run diagnostic. Ligate.
Minipreped inoculations.
3C5: 123.5 ng/mL 280: 1.90 230: 2.86
1K3: 84 ng/mL 280: 1.86 230: 2.58
I0500: 54.7 ng/mL 280: 1.90 230: 2.32
I746908: 234.3 ng/mL 280: 1.85 230: 1.90
Ligation/Promoter characterization (2 10X buffer, 1 ligase):
DNA vector 250-500 ng 400 ng Insert 5-20x of vector 6 ug
Endy’s Promoter F2620 + Superfolded GFP (PCR) + pSB1A2 F2620 + Superfolded GFP (PCR) + pSB3C5 Bad Promoter Testing of I746908 (already in pSB1A2) I746908 + pSB3C5 (10, 5)
Endy’s Promoter T9002 + 1A2 (10, 5) T9002 + 3C5 (10, 5) Bad Promoter I0500 + E0240 + pSB1A2 (5, 5, 5) I0500 + E0240 + pSB3C5 (5, 5, 5)
Promoter/primer design 20 bp
Christopher's Notebook
- Run Gel of Restricted Digested RBS+GFP+Terminators (I746908 partial)
- Gel Extract
- Run Gel of Gel Extracted Digest Products from Friday:
3C5 E/P 2738 bp (buffer 3) F2620 E/S (on pSB1A2) 1061 bp
- I746908 E/P (on psb1A2) 2093 bp-can’t separate
- B0034 X/P (on psb1A2) 12 bp-do NOT run on gel
- J23100 E/S (on psb1A2) 35 bp-do NOT run on gel
Karina's Notebook
Goal: We couldn't extract the terminators because they were too small and ran off the gel. After discussing with Laura and Francisco, we decided to clone GFP and RFP into the plasmid in which the terminators come (pSB1AK3).
Make BSA
- BSA comes as 100x
- made 1 mL of 1x solution by adding 900uL H20 and 100 uL BSA
Cut the GFP and RFP linearized plasmid (prev. cut at S and P) at EcoRI restriction site
- used recipe as described on July 9, 2010
- only used 1 enzyme, therefore used 27 uL H20
- put in waterbath at 10:30 am, and plan to take out after lunch.
Results of diagnostic gel
Observations/Troubleshooting:
- no DNA bands for GFP
- not enough DNA?
- will double DNA concentration during digests
- not enough DNA?
- bands are too large to be RFP
- possible partial digest?
- leave digests overnight
Digests
Digest miniprepped GFP, RFP and Terminators.
- Cut GFP and RFP at E and S
- Cut terminators at E and X
Recipe
- 25 uL DNA
- 1.0 uL enzyme 1
- 1.0 uL enzyme 2
- 5.0 uL NEB Buffer 2
- 5.0 uL BSA (10x)
- 13 uL H20
Total Volume: 50 uL
We used twice as much DNA than the usual protocol because our DNA concentrations from the minipreps were very low.
Prepared 3 reactions for each RSID; ran 6 PCR reactions total.
Ran digests overnight
Francisco's Notebook
- Reran the diagnostic gel (see Karina's Notebook):
- Lessons learned:
- When loading small volumes, use narrow wells.
- When suspecting there are no bands, increase the exposure time to confirm.
Greg's Notebook
- Ran gel with pSB3c5, F2620, I746908
- I746908 didn't work, extracted pSB3c5 and F2620
- Worked on MDV powerpoint presentation
- Ran diagnostic gel of gel extraction results
- pSB3c5 worked, F2620 left only a faint line
- With Alex, miniprepped pSB1k3, I746908, I0500
- Performed nanodrop on miniprep:
| part # | 280 | 230 | concentration (ng/uL) |
| I0500r | 1.87 | 1.28 | 62.5 |
| pSB1k3 | 1.81 | 1.14 | 67.6 |
| I746908 | 1.89 | 1.73 | 175.7 |
- Digested more F2620, pSB3c5, and I0500. Also digested 3 superfolded GFP inserts
7/13 Tuesday
Alex's Notebook
- Streak bacteria
- Back inoculate bacteria 500 ul
Christopher's Notebook
- Run Gels (0.8%) of Restriction Digests:
1M, 2M, 2J (X/P) F2620 (E/S) 3C5 (E/P) I0500 (E/S)
Note: for PCR of I746908: after first gel of PCR product, gel extract, and then do a restriction digest. After restriction digest, do a reaction cleanup; pieces are ready for ligation.
Laura's Notebook
- restreaked plates from 7/1/10 (with Karina)
- for each plate, pick 8 colonies and streak, starting at the center of the "pie piece," and working towards the perimeter of the plate
- plate ID's:
Greg's Notebook
- Made gel, ran digests from yesterday
- Extracted pBAD (I0500) and pSB3c5
- Practiced primer design
- Inventoried my box
7/14 Wednesday
Alex's Notebook
- Order oligos/primers
- Ligate remaining sets
- Streak new bacteria
- Plate old bacteria
- PPT presentation
Laura's Notebook
- plates restreaked yesterday look good- bacteria grew well, some individual colonies
- helped Francisco with minipreps (Promega kit) of : terminator, GFP, RFP
- Francisco set up liquid cultures yesterday afternoon
- Francisco poured, ran 0.8% gel
- yesterday's digestions, with today's minipreps as controls
- gel extraction: (I cut out bands, Francisco and Karina extracted)
| part | tube mass (g) | tube + gel (g) | gel (g) | volume QG (uL) |
| terminator | 1.03 | 1.14 | 0.11 | 330 |
| RFP | 1.11 | 1.18 | 0.07 | 210 |
| GFP | 1.03 | 1.13 | 0.10 | 300 |
Francisco's Notebook
- We've decided to cut out GFP and RFP inserts and ligate them into a linearized terminator plasmid. GFP and RFP inserts are fairly large (~700 bp) and would be easier to gel extract than a terminator insert. The terminator we are using is B1006.
- Mini-prepped GFP, RFP and terminator plasmids. Mini-prep results:
| Part | 260/280 | 260/230 | ng/uL |
|---|---|---|---|
| Term | 1.84 | 0.87 | 62.9 |
| RFP | 1.87 | 1.65 | 235.5 |
| GFP | 1.85 | 1.05 | 121.7 |
- Digested RFP and GFP plasmids with EcoR1 and Spe1; terminator plamids with EcoR1 and Xba1.
- Gel extracted the RFP and GFP inserts and the linearized terminator plasmid.
Greg's Notebook
- Worked on MDV presentation
7/15 Thursday
Alex's Notebook
Check plates; streak new ones; back dilute for a third time. Practice PPT for MDV
7/16 Friday
Recap Meeting Notes
- No recap meeting- presented at MDV this morning.
Alex's Notebook
Due Monday morning: Cloning scheme; check list on wiki for Christina and Drew. Create a timeline for our own use. Deadlines as well.
Francisco's Notebook
- Ran a diagnostic gel of the digestion done on Wednesday
- GFP and RFP plasmids (cut at E, S); Term plasmid (cut at E, X):
- Results: the terminator backbone has a bright band, but the gel extracted GFP and RFP inserts had very faint bands (low concentrations). When we ligate, we may need to use larger volumes of the inserts.
Chris's Notebook
Ligations:
F2620+sfGFP+psb1A2 pBAD+sfGFP+psb1A2 F2620+sfGFP+psb3C5 pBAD+sfGFP+psb3C5
Ligation (20 μl reaction volume):
- DNA vector 250-500 ng (5 ul of vector)
- Insert 5-20x of vector (equal amounts 5 ul of each insert)
- 10x buffer 2 μl
- Ligase 1 μl
- ATP (optional) 1 μl of 10 mM stock (4°C)
- NP water Bring to 20 μl total volume
Ligate at RT for 2 hr (sticky end) or 2 hr (blunt end) Transform 0.5 and 1.0 ul
7/17 Saturday
Alex's Notebook
Check plates again for no growth. Restreaked again.
Run gels for restriction digest products; 120 ul each plasmid type, 240 total. Ten wells minimum total, 4 more lanes for space. Gel extract (partial). Minipreped B0014.
7/18 Sunday
Alex's Notebook
Gel extract (complete). Streak, possible success. Digested B0014 (X/P).
 "
"