Team:Stockholm/3 September 2010
From 2010.igem.org
(New page: {{Stockholm/Top2}} ==Andreas==) |
(→Andreas) |
||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
==Andreas== | ==Andreas== | ||
| + | ===Transfer of m-yCCS into pEX & Cloning of N-CPPs=== | ||
| + | Restreak results from 2/9 revealed four white (positive) clones. These were picked for colony PCR (y5, y6, y8 and y12).<br /> | ||
| + | Also picked four colonies from N-CPP plate from 30/8 (NC1, NC2, NC3 and NC4). | ||
| + | |||
| + | ====Colony PCR==== | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | !colspan="2"|PCR tubes | ||
| + | |- | ||
| + | |dH<sub>2</sub>O | ||
| + | |align="center" width="50"|16.2 | ||
| + | |- | ||
| + | |DreamTaq buffer | ||
| + | |align="center"|2 | ||
| + | |- | ||
| + | |10 mM dNTPs | ||
| + | |align="center"|0.4 | ||
| + | |- | ||
| + | |pEXf | ||
| + | |align="center"|0.4 | ||
| + | |- | ||
| + | |pEXr | ||
| + | |align="center"|0.4 | ||
| + | |- | ||
| + | |DNA | ||
| + | |align="center"|0.5 | ||
| + | |- | ||
| + | |DreamTaq pol. | ||
| + | |align="center"|0.08 | ||
| + | |- | ||
| + | !Total | ||
| + | !20 μl | ||
| + | |} | ||
| + | |||
| + | ====Gel verification==== | ||
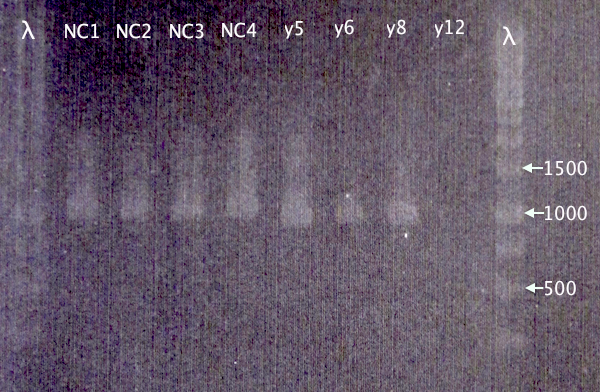
| + | [[image:ColPCR_NCPP_yCCS_3sep.png|200px|right|thumb|'''Gel verification of pSB1C3.N-CPPs and pEX.yCCS colony PCR.'''<br />3 μl λ; 5 μl sample.<br />λ=O'GeneRuler 1 kb DNA ladder]] | ||
| + | 1 % agarose, 80 V | ||
| + | |||
| + | '''Expected bands''' | ||
| + | *pSB1C3.N-CPPs: 365 bp (TAT), 374 bp (LMWP), 395 bp (Tra10) | ||
| + | *pEX.m-yCCS: 963 bp | ||
| + | |||
| + | '''Results'''<br /> | ||
| + | Correct bands for pEX.m-yCCS clones 5, 6 and 8. No band for y12. 5 & 8 will later be selected for plasmid prep.<br /> | ||
| + | Too large bands for all CPP clones to be correct, which is strange. New cloning will be attempted. | ||
| + | |||
| + | ===Cloning of His⋅SOD into pMA=== | ||
| + | ====Plasmid prep==== | ||
| + | ''From 2/9 ON cultures''<br /> | ||
| + | Elution: 50 μl x2 | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | !colspan="3"|DNA concentrations | ||
| + | |- | ||
| + | !Sample | ||
| + | !Conc. [ng/μl] | ||
| + | ! A<sub>260</sub>/A<sub>280</sub> | ||
| + | |- | ||
| + | |pSB1C3.m-yCCS 1 | ||
| + | |align="center"|143.4 | ||
| + | |align="center"|1.91 | ||
| + | |} | ||
| + | |||
| + | ====Sequencing==== | ||
| + | pSB1C3.m-yCCS: ASB0045 A55 | ||
| + | |||
| + | ====Glycerol stock==== | ||
| + | pMA.His⋅SOD 2010-09-03 | ||
| + | |||
| + | ===Cloning of N-CPPs into pSB1C3=== | ||
| + | Re-ligation of 30/8 digestion. | ||
| + | |||
| + | ====Ligation==== | ||
| + | [Dig. pSB1C3 X+A EXTR 1] = 13.72 ng/μl | ||
| + | [Dig. N-CPP X+A] = 66.6 ng/μl | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | !colspan="2"|Ligation mix | ||
| + | |- | ||
| + | |Vector DNA | ||
| + | |align="center" width="50"|6 | ||
| + | |- | ||
| + | |Insert DNA | ||
| + | |align="center"|9 | ||
| + | |- | ||
| + | |5X Rapid Ligation buf. | ||
| + | |align="center"|4 | ||
| + | |- | ||
| + | |dH<sub>2</sub>O | ||
| + | |align="center"|0 | ||
| + | |- | ||
| + | |T4 DNA ligase | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | !Total | ||
| + | !20 μl | ||
| + | |} | ||
| + | |||
| + | ====Transformation==== | ||
| + | Standard transformation procedures. | ||
| + | #: 2 μl ligation mix† | ||
| + | #: 5 μl ligation mix | ||
| + | †: Possibly contaminated due to non-sterile transformation. | ||
| + | |||
| + | ====Gel verification of digestion sample==== | ||
| + | [[image:Gelver dig NCPP 3sep.png|200px|right|thumb|'''Gel verification of undigested and digested (XbaI & AgeI) N-CPP cluster plasmid.'''<br />3 μl λ; 3 μl sample<br />1 kb λ=O'GeneRuler 1 kb DNA ladder; 50 bp λ=GeneRuler 50 bp DNA ladder]] | ||
| + | *NCPP: undigested N-CPP cluster plasmid | ||
| + | *Dig NCPP: digested N-CPP cluster plasmid, XbaI and AgeI | ||
| + | |||
| + | 1 % agarose, 100 V | ||
| + | |||
| + | '''Results'''<br /> | ||
| + | Difficult to interpret results, since the source plasmid size is unknown. However, it looks like the digested sample presents a shorter fragment compared to the undigested. On the other hand, it also looks like the size difference is too big to have resulted from just excising the ≈300 bp N-CPP cluster.<br /> | ||
| + | Also strange is that it looks like there are two bands present in the undigested sample. | ||
Revision as of 11:12, 6 September 2010
Contents |
Andreas
Transfer of m-yCCS into pEX & Cloning of N-CPPs
Restreak results from 2/9 revealed four white (positive) clones. These were picked for colony PCR (y5, y6, y8 and y12).
Also picked four colonies from N-CPP plate from 30/8 (NC1, NC2, NC3 and NC4).
Colony PCR
| PCR tubes | |
|---|---|
| dH2O | 16.2 |
| DreamTaq buffer | 2 |
| 10 mM dNTPs | 0.4 |
| pEXf | 0.4 |
| pEXr | 0.4 |
| DNA | 0.5 |
| DreamTaq pol. | 0.08 |
| Total | 20 μl |
Gel verification
1 % agarose, 80 V
Expected bands
- pSB1C3.N-CPPs: 365 bp (TAT), 374 bp (LMWP), 395 bp (Tra10)
- pEX.m-yCCS: 963 bp
Results
Correct bands for pEX.m-yCCS clones 5, 6 and 8. No band for y12. 5 & 8 will later be selected for plasmid prep.
Too large bands for all CPP clones to be correct, which is strange. New cloning will be attempted.
Cloning of His⋅SOD into pMA
Plasmid prep
From 2/9 ON cultures
Elution: 50 μl x2
| DNA concentrations | ||
|---|---|---|
| Sample | Conc. [ng/μl] | A260/A280 |
| pSB1C3.m-yCCS 1 | 143.4 | 1.91 |
Sequencing
pSB1C3.m-yCCS: ASB0045 A55
Glycerol stock
pMA.His⋅SOD 2010-09-03
Cloning of N-CPPs into pSB1C3
Re-ligation of 30/8 digestion.
Ligation
[Dig. pSB1C3 X+A EXTR 1] = 13.72 ng/μl [Dig. N-CPP X+A] = 66.6 ng/μl
| Ligation mix | |
|---|---|
| Vector DNA | 6 |
| Insert DNA | 9 |
| 5X Rapid Ligation buf. | 4 |
| dH2O | 0 |
| T4 DNA ligase | 1 |
| Total | 20 μl |
Transformation
Standard transformation procedures.
- 2 μl ligation mix†
- 5 μl ligation mix
†: Possibly contaminated due to non-sterile transformation.
Gel verification of digestion sample
- NCPP: undigested N-CPP cluster plasmid
- Dig NCPP: digested N-CPP cluster plasmid, XbaI and AgeI
1 % agarose, 100 V
Results
Difficult to interpret results, since the source plasmid size is unknown. However, it looks like the digested sample presents a shorter fragment compared to the undigested. On the other hand, it also looks like the size difference is too big to have resulted from just excising the ≈300 bp N-CPP cluster.
Also strange is that it looks like there are two bands present in the undigested sample.
 "
"