Team:UNIPV-Pavia/Calendar/June/settimana4
From 2010.igem.org
(→June, 25th) |
|||
| (45 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
<td valign="top"> | <td valign="top"> | ||
<table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> | <table border="0" align="center" width="100%"><tr><td align="justify" valign="top" style="padding:20px"> | ||
| - | |||
| + | <table class="menu" border="0" width="100%"> | ||
| + | <tr> | ||
| + | <td align="center" style="padding:0; height:20px"> | ||
| + | [[Team:UNIPV-Pavia/Calendar/June/settimana2|Week 2]] | ||
| + | </td> | ||
| + | <td align="center"> | ||
| + | [[Team:UNIPV-Pavia/Calendar/June/settimana3|Week 3]] | ||
| + | </td> | ||
| + | <td align="center"> | ||
| + | [[Team:UNIPV-Pavia/Calendar/June/settimana4|Week 4]] | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | |||
| + | <html><p align="center"><font size="4"><b>JUNE: WEEK 4</b></font></p></html><hr><br> | ||
| + | <html><a name="indice"/></html> | ||
==June, 21st== | ==June, 21st== | ||
| + | |||
| + | Planning of the activity of the week. | ||
| + | Freezer cleaning: for each ligation, we chose the correct clone and stored it in the ''iGEM 2010 ligations'' box. All these cloned were gel screened and sequenced and are correct! | ||
| + | The colonies we chose are: | ||
| + | <table align='center' width='30%' border='1'> | ||
| + | <tr align='center'><td>'''colony chosen'''</td> | ||
| + | <td>'''ligation name'''</td></tr> | ||
| + | <tr align='center'><td>I0-2</td><td>I0</td></tr> | ||
| + | <tr align='center'><td>I1-2</td><td>I1</td></tr> | ||
| + | <tr align='center'><td>I2-1</td><td>I2</td></tr> | ||
| + | <tr align='center'><td>I3-1</td><td>I3</td></tr> | ||
| + | <tr align='center'><td>I4-2</td><td>I4</td></tr> | ||
| + | <tr align='center'><td>I5-1</td><td>I5</td></tr> | ||
| + | <tr align='center'><td>I6-2</td><td>I6</td></tr> | ||
| + | </table> | ||
| + | |||
| + | Other colonies resulted positives at the screening but were NOT sequenced and are stored in the ''iGEM 2010 cemetery'' box. These clones are: I1-1, I2-2, I4-1 and I6-3. | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==June, 22nd== | ==June, 22nd== | ||
| + | |||
| + | |||
| + | Inoculum of I6, <partinfo>BBa_J23118</partinfo>, <partinfo>BBa_J23110</partinfo>, <partinfo>BBa_J23114</partinfo>, <partinfo>BBa_J23116</partinfo> from glycerol stock in 5ml LB+Amp. Cultures were grown ON at 37°C 220rpm. | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==June, 23rd== | ==June, 23rd== | ||
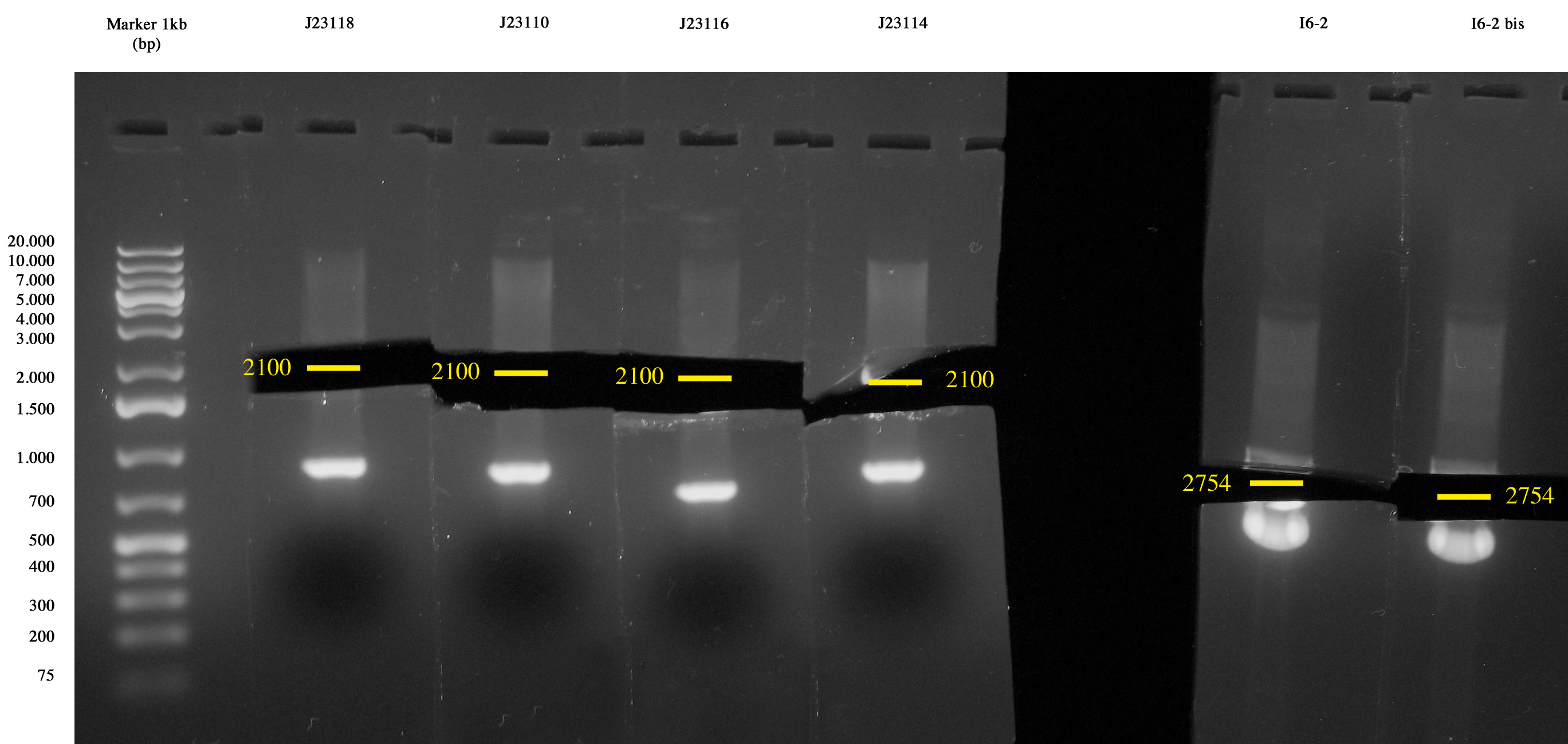
| + | |||
| + | Cultures incubated ON grew all. Plasmids were extracted with MiniPrep. | ||
| + | |||
| + | |||
| + | [[Image:Unipv_digestion_J231xx_I6.jpg|300px|thumb|center|J231xx and I6 digestion]] | ||
| + | |||
| + | After MiniPrep, purified DNA was quantified with NanoDrop. | ||
| + | |||
| + | {| border="1" align='center' | ||
| + | | I6 || 193 ng/ul | ||
| + | |- | ||
| + | | <partinfo>BBa_J23118</partinfo> || 107,9 ng/ul | ||
| + | |- | ||
| + | | <partinfo>BBa_J23110</partinfo> || 83 ng/ul | ||
| + | |- | ||
| + | | <partinfo>BBa_J23114</partinfo> || 89,8 ng/ul | ||
| + | |- | ||
| + | | <partinfo>BBa_J23116</partinfo> || 82,7 ng/ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Digestion of: | ||
| + | |||
| + | {| border="1" align='center' | ||
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1'' || ''Enzyme 2'' || ''Buffer H'' | ||
| + | |- | ||
| + | | I6 || Insert || 25 || 9,3 || 11,2 || 1 XbaI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | I6-bis || Insert || 25 ||9,3 || 11,2 || 1 XbaI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23118</partinfo> || Vector || 25 || 9,3 || 11,2 || 1 SpeI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23110</partinfo> || Vector || 25 ||12 || 8,5 || 1 SpeI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23114</partinfo> || Vector || 25 || 11,2 || 9,3 || 1 SpeI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | <partinfo>BBa_J23116</partinfo> || Vector || 25 || 12,1 || 8,4 || 1 SpeI || 1 PstI || 2,5 | ||
| + | |} | ||
| + | |||
| + | Digestions were incubated at 37°C for 3 hours, gel run and gel-extracted. | ||
| + | |||
| + | Ligations were performed ON at 16°C: | ||
| + | |||
| + | *I7: <partinfo>BBa_J23118</partinfo> (S-P)+ I6 (X-P) | ||
| + | *I8: <partinfo>BBa_J23110</partinfo> (S-P)+ I6 (X-P) | ||
| + | *I9: <partinfo>BBa_J23114</partinfo> (S-P)+ I6 (X-P) | ||
| + | *I10: <partinfo>BBa_J23116</partinfo> (S-P)+ I6 (X-P) | ||
| + | |||
| + | NanoDrop quantifications were not reliable, so every ligation was performed with 3ul of vector, 2ul of insert, 3ul of ddH20, 1ul of T4 ligase and 1ul of T4 ligase buffer. | ||
| + | |||
| + | These four promoters from Anderson Promoters Collection were chose for their ''strength'', measured in Arbitrary Units: | ||
| + | |||
| + | {| border='1' align='center' | ||
| + | || '''Part''' || '''RFP (a.u.)''' | ||
| + | |- | ||
| + | ||<partinfo>BBa_J23118</partinfo>||1429 | ||
| + | |- | ||
| + | ||<partinfo>BBa_J23110</partinfo>|| 844 | ||
| + | |- | ||
| + | ||<partinfo>BBa_J23114</partinfo>|| 256 | ||
| + | |- | ||
| + | ||<partinfo>BBa_J23116</partinfo>|| 396 | ||
| + | |- | ||
| + | |} | ||
| + | Soon quantitative tests will be performed to quantify each promoter's strength in terms of RPU. | ||
| + | |||
| + | Today we received 2 stabs from iGEM HQ for <partinfo>BBa_K208001</partinfo> (one for each registry location of this part), the Silver-fusion compatible BioBrick part that codes for phasin. This part is in a pSB3K3 plasmid. Bacteria were streaked on LB+Kan (50 ug/ml) agar plates. Cultures were named PhaP-1 and PhaP-2. Plates were incubated ON at 37°C. | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==June, 24th== | ==June, 24th== | ||
| + | |||
| + | PhaP-1 and PhaP-2 plates showed colonies!! | ||
| + | A colony from each plate was picked and used to infect 1ml LB+Kan. Liquid cultures of PhaP-1 and PhaP-2 were grown for 6 hours at 37°C 220 rpm and then used to prepare glycerol stocks. Remaining liquid cultures were re-filled with 5 ml LB+Kan an grown ON at 37°C 220 rpm for tomorrow MiniPrep. | ||
| + | Glycerol stocks of PhaP-1 and PhaP-2 are stored at -80°C in the ''iGEM 2010 Registry'' box. | ||
| + | |||
| + | Ligations were heated at 65°C for 5 minutes to inactivate ligase ad then tranformed in E. coli TOP10 home made competent cells. Plates were incubated ON at 37°C (we decided to incubate plates 5 hours longer than usual, because cell growth was already slower for I6). | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==June, 25th== | ==June, 25th== | ||
| - | + | All plates showed colonies after 19-hour growth at 37°C!! | |
| + | *I7: small green colonies with satellite colonies were observed. Negative colonies are present and can be easily distinguished, because they are red. | ||
| + | *I8: green colonies were observed, few negative colonies are pink. | ||
| + | *I9: big colonies were observed, some of them are surrounded by satellite colonies. | ||
| + | *I10: big colonies were observed, some light pink colonies are present, surrounded by satellite colonies. | ||
| + | These results are encouraging, because they are consistent with our expectations: ligations provide a high metabolic burden to the cell, so growth is slower. ''Positive'' green colonies are smaller and not surrounded by satellite colonies. ''Negative'' colonies are bigger and surrounded by satellite colonies, confirming that they had a faster growth than our ligations! | ||
| - | <td width= | + | Two colonies from each plate were picked and incubated in 1ml LB+Amp. |
| + | <table> | ||
| + | <tr><td width='50%'> | ||
| + | <table border='1' align='center'> | ||
| + | <tr align='center'><td><b>culture</b></td></tr> | ||
| + | <tr align'center'><td>I7-1</td></tr> | ||
| + | <tr align'center'><td>I7-2</td></tr> | ||
| + | <tr align'center'><td>I8-1</td></tr> | ||
| + | <tr align'center'><td>I8-2</td></tr> | ||
| + | <tr align'center'><td>I9-1</td></tr> | ||
| + | <tr align'center'><td>I9-2</td></tr> | ||
| + | <tr align'center'><td>I10-1</td></tr> | ||
| + | <tr align'center'><td>I10-2</td></tr> | ||
| - | <table | + | </table> |
| - | + | ||
| - | + | </td><td> | |
| - | </td | + | Cultures were grown for 6 hours at 37°C 220 rpm, then glycerol stocks were prepared for each culture. Glycerol stocks are stored at -80°C in ''iGEM 2010 ligations'' box. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</td></tr> | </td></tr> | ||
</table> | </table> | ||
| + | PhaP-1 and PhaP-2 cultures were grown. MiniPre was performed, with the following NanoDrop quantifications: | ||
| - | </td> | + | {| border="1" |
| + | | PhaP-1 || 9,6 ng/ul | ||
| + | |- | ||
| + | | PhaP-2 || 4,3 ng/ul | ||
| + | |} | ||
| + | |||
| + | DNA quantification was poor, so we decided to perform digestion screening and to repeat Miniprep next week for sequencing. | ||
| + | Digestion of: | ||
| + | |||
| + | {| border="1" | ||
| + | | ''Culture'' || ''Kind'' || ''Final reaction volume (ul) '' || ''DNA (ul)'' || ''H20 (ul)'' || ''Enzyme 1'' || ''Enzyme 2'' || ''Buffer H'' | ||
| + | |- | ||
| + | | PhaP-1 || Screening || 25 || 20,5 || 0 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |- | ||
| + | | PhaP-2 || Screening || 25 || 20,5 || 0 || 1 EcoRI || 1 PstI || 2,5 | ||
| + | |} | ||
| + | |||
| + | Digestion was performed for 2 hours at 37°C. DNA was then gel run for screening. Both cultures were positive! Sequencing will be performed next week! | ||
| + | |||
| + | [[Image:Unipv_screening_PhaP_1_2.jpg|200px|thumb|center|Screening of PhaP-1 and PhaP-2]] | ||
| + | |||
| + | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| + | |||
| + | <!-- table previous next week --> | ||
| + | <br><br> | ||
| + | <table border="0" width="100%" class="menu"> | ||
| + | <tr> | ||
| + | <td align="left">[[Team:UNIPV-Pavia/Calendar/June/settimana3| Previous week]]</td> | ||
| + | <td align="right">[[Team:UNIPV-Pavia/Calendar/July/settimana1| Next week]]</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | <!-- fine table previous next week --> | ||
| + | </td> | ||
| + | |||
| + | <td width="15%" align="right" valign="top"> | ||
| + | |||
| + | {{UNIPV-Pavia/menu_mesi}} | ||
</td> | </td> | ||
| + | |||
| + | </tr> | ||
| + | </table> | ||
Latest revision as of 08:00, 31 August 2010
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"