Team:Stockholm/10 August 2010
From 2010.igem.org
NinaSchiller (Talk | contribs) (→Transformation of protein A (ZZ domain) in bank vector C) |
(→Mimmi) |
||
| Line 195: | Line 195: | ||
| 10 | | 10 | ||
|} | |} | ||
| + | |||
| + | |||
| + | |||
| + | === Gels === | ||
| + | |||
| + | ==== MITF ==== | ||
| + | ::'''verification''' | ||
| + | |||
| + | *Nothing...! | ||
| + | |||
| + | |||
| + | ==== yCCS and RFP ==== | ||
| + | ::'''verification''' | ||
| + | |||
| + | *See Andreas notes | ||
| + | |||
| + | |||
| + | ==== yCCS and SOD ==== | ||
| + | ::'''site-directed''' | ||
| + | |||
| + | [[Image:2010-08-10_yCCS_+_SOD_site_directed_mutagsenesis.jpg|200px|thumb|left|]] | ||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | yCCS | ||
| + | |- | ||
| + | | 3 | ||
| + | | blank | ||
| + | |- | ||
| + | | 4 | ||
| + | | plasmid control | ||
| + | |- | ||
| + | | 5 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 6 | ||
| + | | SOD | ||
| + | |- | ||
| + | | 7 | ||
| + | | blank | ||
| + | |- | ||
| + | | 8 | ||
| + | | plasmid control | ||
| + | |} | ||
| + | |||
| + | |||
---- | ---- | ||
| + | |||
==Nina== | ==Nina== | ||
Revision as of 11:25, 18 August 2010
Contents |
Mimmi
yCCS and SOD
redoing the Site-Directed Mutagenesis
| Mix | (µl) | X2 X2 | yCCS A | SOD A | conditions | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| sH2O | 40 | 80 | yCCS_mut_F | SOD_mut_F | time | °C | ||||
| dNTPs | 1 | 2 | yCCS_mut_R | SOD_mut_R | 30s | 95 | ||||
| F primer | 1 | 2 | pSB1C3 | pSB1C3 | 30s | 95 | ) | |||
| R primer | 1 | 2 | ~3.5kb | ~3.2kb | 30s | 55 | > 22 cycles | |||
| DNA | 1 | 2X1 | 4m | 68 | ) | |||||
| Pfu buffer | 5 | 10 | oo | 10 | ||||||
| Pfu turbo | 1 | 2 | ||||||||
| tot | 50 | |||||||||
SOD Primers SOD_mut_F 262.55µl H2O --> 100µM SOD_mut_R 372.64µl H2O --> 100µM SOD_mut_F 3µl + 24.15µl H2O --> 125ng/µl SOD_mut_R 3µl + 24.42µl H2O --> 125ng/µl
MITF
Amplifying
| Mix phusion | (µl) | /4 | Mix Pfu turbo | (µl) | /4 | Primers | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| sH2O | 67 | sH2O | 77 | MITF_F | ||||||
| F primer | 5 | F primer | 5 | MITF_R | ||||||
| R primer | 5 | R primer | 5 | |||||||
| dNTP | 2 | dNTP | 2 | conditions | ||||||
| 5X buffer | 20 | 10X buffer | 10 | time | °C | |||||
| Phusion pol. | 1 | Pfu pol. | 1 | 2m | 98 | |||||
| tot | 100 | 25 | tot | 100 | 25 | 30s | 98 | ) | ||
| 30s | 40-55 | > 5 cycles | ||||||||
| 1m30s | 72 | ) | ||||||||
| 30s | 98 | ) | ||||||||
| 30s | 65 | > 25 cycles | ||||||||
| 1m30s | 72 | ) | ||||||||
| 10m | 72 | |||||||||
| oo | 10 | |||||||||
Gels
MITF
- verification
- Nothing...!
yCCS and RFP
- verification
- See Andreas notes
yCCS and SOD
- site-directed
| well | sample |
|---|---|
| 1 | ladder |
| 2 | yCCS |
| 3 | blank |
| 4 | plasmid control |
| 5 | ladder |
| 6 | SOD |
| 7 | blank |
| 8 | plasmid control |
Nina
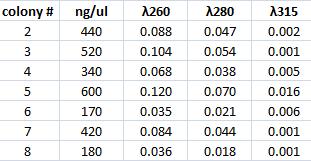
Mini prep of Tyrosinase
I performed a mini prep on the six samples of falcon tubes containing inoculated tyrosinase in its original vector that have been site directed mutagenesis. The colonies are: #2, 3, 4, 5, 6, ,7 & 8. The method was according to the procedure described under Protocols.
Spectophotometer:
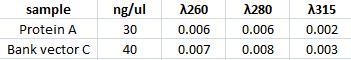
Measuring concentration of protein A (ZZ domain) and IgG protease in bank vector C
I measured the complately digested protein A ZZ domain (PCR product) and the bank vector C that before the digestion contained the IgG protease gene.
Spectophotometer:
Ligation of protein A (ZZ domain) with bank vector C
I ligated protein A ZZ domian into the digested bank vector C to be able to make fusion proteins with IgG protease and CPP but also to send in this part to iGEM hq since they want the genes to be sent in this vector.
The vector concentraion should preferably be 25 ng/ul when performing ligation with a gene.
I used:
- bank vector C 0.5 ul
- protein A gene 3 ul
- quick ligase 1 ul
- quicke ligase buffer 2X 3 ul
Incubated in RT 15 min
Transformation of protein A (ZZ domain) in bank vector C
I transformed the ligated protein A (ZZ domain) in bank vector C in 100 ul Top 10 cells.
The transformation method is according to the procedure decribed in protocols. However in the step 1 I thawed for 15 min instead of 10 min. In step 2 I added 3 ul of DNA, in step 6 I used LB instead of SOC and finally in step 10 I spread 50 ul of sample on a chloramphenicol plate.
Andreas
Cloning of IgG protease
Continued from 9/8 ON plates
Colony PCR
Four clones (A, B, C, D) were picked for colony PCR. Positive control: pSB1C3.BBa_J04450 plasmid. Procedures according to protocol; 1:40 elongation time.
Gel verification
1 % agarose, 90 V, 40 min
Results: Weak and very irregularly sized bands. Probably something wrong with the ligation mixture. New IgGp digestion will be made.
New IgG protease cloning
Digestion
Used pSB1A3.IgGp at a conc. of 139 ng/μl.
| 10X FD buffer | 2 μl |
| dH2O | 3 μl |
| 2 μg DNA | 14 μl |
| FD SpeI | 0.5 μl |
| FD EcoRI | 0.5 μl |
| 20 μl |
Incubation: 37 °C, 30 min
Ligation
Used pre-digested (EcoRI & SpeI) pSB1C3 vector from 3/8.
| pSB1C3 vector | 2.5 μl |
| IgGp insert | 12.5 μl † |
| 5X rapid ligation buffer | 4 μl |
| T4 DNA ligase | 1 μl |
† Since I used the ligation protocol from 3/8, I mistakenly calculated that 12.5:2.5 would give a 5:1 insert:vector ratio. However, due to the new digestion volume, this actually led to a 12.5:1 ratio.
Transformation
Transformation into 100 μl comptetent Top10.
- Quick-transformation
- 3 μl ligation mix. Incubation 5 min on ice.
- Heat-shock 30 sec in 42 °C. Cells cooled on ice.
- 100 μl plated onto Cm 25 plate.
- Standard transformation
- 3 μl ligation mix.
- Standard protocol transformation procedures.
Plates incubated ON in 37 °C.
 "
"