Team:TU Delft/22 July 2010 content
From 2010.igem.org
(New page: ==Alkane degradation== There were some colonies on [https://2010.igem.org/Team:TU_Delft#/blog?blog=19_July_2010 Tuesday's] plates! We had left the plates @ 37°C yesterday after having seen...) |
(→Method 4) |
||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | =Lab work= | ||
| + | |||
==Alkane degradation== | ==Alkane degradation== | ||
| - | There were some colonies on [https://2010.igem.org/Team:TU_Delft#/blog | + | There were some colonies on [https://2010.igem.org/Team:TU_Delft#page=pages/blog&blog=19_July_2010 Tuesday's] plates! We had left the plates @ 37°C yesterday after having seen that there were no colonies. When checking this morning on all plates (except the negative control) there were a few colonies! (2-50 colonies). Chances are it's not what we're looking for, but maybe they are good transformants... to check we will do a [[Team:TU_Delft/protocols/colony PCR|colony PCR]]. |
| + | |||
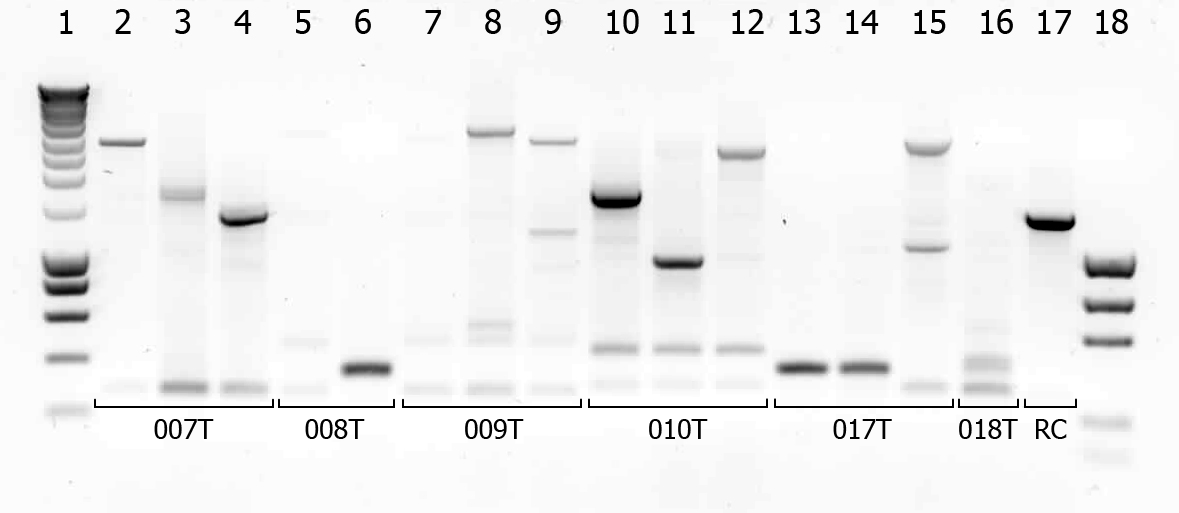
| + | [[Image:TUDelft_scPCR_22-07.png|550px|thumb|left|1% Agarose gel of colony PCR. Gel runned at 100V for 1 hour. Of all samples 5 μL + 1 μL loadingbuffer was loaded. 5 μL was loaded of marker.]] | ||
| + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Description''' | ||
| + | |'''Expected length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |- | ||
| + | |1 | ||
| + | |[https://2010.igem.org/Image:TU_Delft_SmartLadder.jpg SmartLadder] | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |n/a | ||
| + | |- | ||
| + | |2 | ||
| + | |Transformant #1 of ligation mix 007T | ||
| + | |1616 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |Transformant #2 of ligation mix 007T | ||
| + | |1616 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |Transformant #3 of ligation mix 007T | ||
| + | |1616 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |5 | ||
| + | |Transformant #1 of ligation mix 008T | ||
| + | |551 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |6 | ||
| + | |Transformant #2 of ligation mix 008T | ||
| + | |551 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |7 | ||
| + | |Transformant #1 of ligation mix 009T | ||
| + | |551 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |8 | ||
| + | |Transformant #1 of ligation mix 009T | ||
| + | |560 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |9 | ||
| + | |Transformant #1 of ligation mix 009T | ||
| + | |560 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |10 | ||
| + | |Transformant #1 of ligation mix 010T | ||
| + | |1657 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |11 | ||
| + | |Transformant #1 of ligation mix 010T | ||
| + | |1657 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |12 | ||
| + | |Transformant #1 of ligation mix 010T | ||
| + | |1657 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |13 | ||
| + | |Transformant #1 of ligation mix 017T | ||
| + | |1130 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |14 | ||
| + | |Transformant #1 of ligation mix 017T | ||
| + | |1130 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |15 | ||
| + | |Transformant #1 of ligation mix 017T | ||
| + | |1130 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |16 | ||
| + | |Transformant #1 of ligation mix 018T | ||
| + | |1874 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |- | ||
| + | |17 | ||
| + | |Transformant #1 of Red colony | ||
| + | |1360 | ||
| + | |G00100 + G00101 | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | A number of colonies look promising! To check if they really are the BioBricks we want, tomorrow we will do a plasmid isolation with the cultures of lane 2, 4, 6, 7, 9 and 14. We will cut the isolated plasmids with various restriction enzymes and analyze the digestion products on gel. | ||
| + | |||
| + | ==Characterisation of Anderson RBS sequences== | ||
| + | |||
| + | ====Assembly of reference construct==== | ||
| + | |||
| + | =====Method 3===== | ||
| + | |||
| + | The Amp plates contained very few transformants. These were taken for [https://2010.igem.org/Team:TU_Delft#page=protocols/colony_PCR single colony PCR] and run on [https://2010.igem.org/Team:TU_Delft#page=protocols/agarose_gel 1% agarose gel]: | ||
| + | |||
| + | [[Image:TU_Delft_SCpcr_22-07_(pcr_gen).png|450px|thumb|left|1% agarose of colony PCR. Gel run at 100V for 1 hour. Of all samples 5 µL was loaded with 1 µL loadingbuffer. 5 µL was loaded of marker]] | ||
| + | |||
| + | Lane description: | ||
| + | {|style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Description''' | ||
| + | |'''Expected Length (bp)''' | ||
| + | |'''Primers''' | ||
| + | |'''Status''' | ||
| + | |'''Remarks''' | ||
| + | |- | ||
| + | |1 | ||
| + | |SmartLadder | ||
| + | |Varies | ||
| + | |n/a | ||
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |Transformant #1 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | ||
| + | |1157 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>?</font> | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |Transformant #2 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | ||
| + | |1157 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>?</font> | ||
| + | | | ||
| + | |- | ||
| + | |4 | ||
| + | |Transformant #3 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | ||
| + | |1157 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>?</font> | ||
| + | | | ||
| + | |- | ||
| + | |5 | ||
| + | |Transformant #4 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | ||
| + | |1157 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>?</font> | ||
| + | | | ||
| + | |- | ||
| + | |6 | ||
| + | |Transformant #5 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | ||
| + | |1157 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>?</font> | ||
| + | | | ||
| + | |- | ||
| + | |7 | ||
| + | |Transformant #6 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | ||
| + | |1157 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=red>?</font> | ||
| + | | | ||
| + | |- | ||
| + | |8 | ||
| + | |PCR product of J23100 in pSB1A2 | ||
| + | |1142 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
| + | |- | ||
| + | |9 | ||
| + | |PCR product of E0240 in pSB1A2 | ||
| + | |1114 | ||
| + | |G00100 + G00101 | ||
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
| + | |- | ||
| + | |10 | ||
| + | |BioRad EZ Load | ||
| + | |Varies | ||
| + | |n/a | ||
| + | |<font color=limegreen>✓</font> | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Due to the very short expected difference in bp between the constructs, the gel could not be conclusive. However, the transformants di not fluoresce when screened with UV excitation. | ||
| + | |||
| + | =====Method 4===== | ||
| + | |||
| + | After having stored yesterday's Amp plates in the fridge it was decided to screen once more for GFP fluorescence. This time multiple fluorescing colonies were found under UV excitation, indicating that the reference construct J23100-E0240 had been created. Three of the colonies were brought over to 5 mL of LB+AMP for over night culturing and characterization. | ||
Latest revision as of 21:56, 16 August 2010
Contents |
Lab work
Alkane degradation
There were some colonies on Tuesday's plates! We had left the plates @ 37°C yesterday after having seen that there were no colonies. When checking this morning on all plates (except the negative control) there were a few colonies! (2-50 colonies). Chances are it's not what we're looking for, but maybe they are good transformants... to check we will do a colony PCR.
| # | Description | Expected length (bp) | Primers | Status |
| 1 | SmartLadder | n/a | n/a | n/a |
| 2 | Transformant #1 of ligation mix 007T | 1616 | G00100 + G00101 | |
| 3 | Transformant #2 of ligation mix 007T | 1616 | G00100 + G00101 | |
| 4 | Transformant #3 of ligation mix 007T | 1616 | G00100 + G00101 | |
| 5 | Transformant #1 of ligation mix 008T | 551 | G00100 + G00101 | |
| 6 | Transformant #2 of ligation mix 008T | 551 | G00100 + G00101 | |
| 7 | Transformant #1 of ligation mix 009T | 551 | G00100 + G00101 | |
| 8 | Transformant #1 of ligation mix 009T | 560 | G00100 + G00101 | |
| 9 | Transformant #1 of ligation mix 009T | 560 | G00100 + G00101 | |
| 10 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | |
| 11 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | |
| 12 | Transformant #1 of ligation mix 010T | 1657 | G00100 + G00101 | |
| 13 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | |
| 14 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | |
| 15 | Transformant #1 of ligation mix 017T | 1130 | G00100 + G00101 | |
| 16 | Transformant #1 of ligation mix 018T | 1874 | G00100 + G00101 | |
| 17 | Transformant #1 of Red colony | 1360 | G00100 + G00101 |
A number of colonies look promising! To check if they really are the BioBricks we want, tomorrow we will do a plasmid isolation with the cultures of lane 2, 4, 6, 7, 9 and 14. We will cut the isolated plasmids with various restriction enzymes and analyze the digestion products on gel.
Characterisation of Anderson RBS sequences
Assembly of reference construct
Method 3
The Amp plates contained very few transformants. These were taken for single colony PCR and run on 1% agarose gel:
Lane description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| 1 | SmartLadder | Varies | n/a | ✓ | |
| 2 | Transformant #1 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | 1157 | G00100 + G00101 | ? | |
| 3 | Transformant #2 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | 1157 | G00100 + G00101 | ? | |
| 4 | Transformant #3 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | 1157 | G00100 + G00101 | ? | |
| 5 | Transformant #4 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | 1157 | G00100 + G00101 | ? | |
| 6 | Transformant #5 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | 1157 | G00100 + G00101 | ? | |
| 7 | Transformant #6 of ligation mix: S-J23100-pSB1A2-P + X-E0240-P (PCR) | 1157 | G00100 + G00101 | ? | |
| 8 | PCR product of J23100 in pSB1A2 | 1142 | G00100 + G00101 | ✓ | |
| 9 | PCR product of E0240 in pSB1A2 | 1114 | G00100 + G00101 | ✓ | |
| 10 | BioRad EZ Load | Varies | n/a | ✓ |
Due to the very short expected difference in bp between the constructs, the gel could not be conclusive. However, the transformants di not fluoresce when screened with UV excitation.
Method 4
After having stored yesterday's Amp plates in the fridge it was decided to screen once more for GFP fluorescence. This time multiple fluorescing colonies were found under UV excitation, indicating that the reference construct J23100-E0240 had been created. Three of the colonies were brought over to 5 mL of LB+AMP for over night culturing and characterization.
 "
"